5GR0
 
 | |
6Z39
 
 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY functionalised epoxide activity based probe | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[4-[4-[(12~{R})-2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9-trien-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
6DT3
 
 | | 1.2 Angstrom Resolution Crystal Structure of Nucleoside Triphosphatase NudI from Klebsiella pneumoniae in Complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nucleoside triphosphatase NudI | | Authors: | Minasov, G, Shuvalova, L, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
7V37
 
 | | Crystal structure of apo-NP exonuclease | | Descriptor: | Nucleoprotein, ZINC ION | | Authors: | Hsiao, Y.Y, Huang, K.W. | | Deposit date: | 2021-08-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Targeted Covalent Inhibitors Allosterically Deactivate the DEDDh Lassa Fever Virus NP Exonuclease from Alternative Distal Sites.
Jacs Au, 1, 2021
|
|
5ZCX
 
 | | Structure of T20/N39 | | Descriptor: | Envelope glycoprotein, Envelope glycoprotein gp160 | | Authors: | Zhang, X.J, Ding, X.H. | | Deposit date: | 2018-02-21 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural and functional characterization of HIV-1 cell fusion inhibitor T20.
AIDS, 33, 2019
|
|
6H4Z
 
 | | Crystal structure of human KDM5B in complex with compound 16a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-[4-(3-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
5H9W
 
 | | Crystal structure of Regnase PIN domain, form II | | Descriptor: | Ribonuclease ZC3H12A, SODIUM ION | | Authors: | Yokogawa, M, Tsushima, T, Adachi, W, Noda, N.N, Inagaki, F. | | Deposit date: | 2015-12-29 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions
Sci Rep, 6, 2016
|
|
6BKY
 
 | | Novel Binding Modes of Inhibition of Wild-Type IDH1: Allosteric Inhibition with Cmpd2 | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 4,5,6,7-TETRABROMO-1H,3H-BENZIMIDAZOL-2-ONE, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2017-11-09 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Modes of Inhibition of Wild-Type Isocitrate Dehydrogenase 1 (IDH1): Direct Covalent Modification of His315.
J. Med. Chem., 61, 2018
|
|
7YA4
 
 | |
6BUM
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica | | Descriptor: | 1,3-PROPANDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shi, K, Cho, S, Seffernick, J.L, Bera, A, Wackett, L.P, Aihara, H. | | Deposit date: | 2017-12-11 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
9IHQ
 
 | |
9IHP
 
 | |
3MJY
 
 | | Crystal structure of dihydroorotate dehydrogenase from Leishmania major in complex with 5-Aminoorotic acid | | Descriptor: | 5-amino-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, Dihydroorotate dehydrogenase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Rocha, J.R, Cheleski, J, Montanari, C.A, Nonato, M.C. | | Deposit date: | 2010-04-13 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Novel insights for dihydroorotate dehydrogenase class 1A inhibitors discovery.
Eur.J.Med.Chem., 45, 2010
|
|
6JX2
 
 | | Crystal structure of Ketol-acid reductoisomerase from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Lee, D, Hong, J, Kim, K.-J. | | Deposit date: | 2019-04-22 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure and Biochemical Characterization of Ketol-Acid Reductoisomerase fromCorynebacterium glutamicum.
J.Agric.Food Chem., 67, 2019
|
|
6SNP
 
 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | MAGNESIUM ION, Phosphoglucomutase-1 | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
6FN6
 
 | | Modifying region (DH-ER-KR) of an insect fatty acid synthase (FAS) | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Fatty acid synthase 1, ... | | Authors: | Benning, F.M.C, Bukhari, H.S.T, Jakob, R.P, Maier, T. | | Deposit date: | 2018-02-02 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Modifying region (DH-ER-KR) of an insect fatty acid synthase (FAS)
To Be Published
|
|
6RFR
 
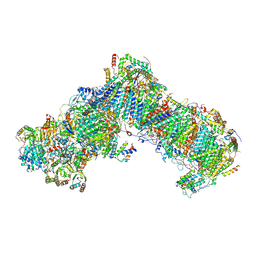 | | Cryo-EM structure of respiratory complex I from Yarrowia lipolytica at 3.2 A resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
9IHO
 
 | |
8PPN
 
 | |
5I4T
 
 | |
6Y6S
 
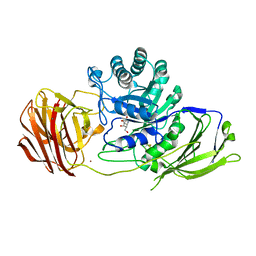 | | Mouse Galactocerebrosidase complexed with galacto-noeurostegine GNS at pH 6.8 | | Descriptor: | (1~{R},2~{S},3~{S},4~{R},5~{R})-4-(hydroxymethyl)-8-azabicyclo[3.2.1]octane-1,2,3-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Deane, J.E, McLoughlin, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Bicyclic Form of galacto -Noeurostegine Is a Potent Inhibitor of beta-Galactocerebrosidase.
Acs Med.Chem.Lett., 12, 2021
|
|
6I7Q
 
 | |
5C77
 
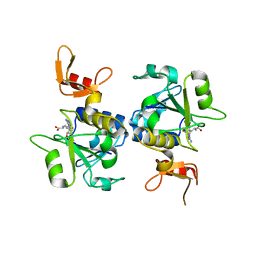 | | A novel protein arginine methyltransferase | | Descriptor: | Protein arginine N-methyltransferase SFM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lv, F, Zhang, T, Ding, J. | | Deposit date: | 2015-06-24 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for Sfm1 functioning as a protein arginine methyltransferase.
Cell Discov, 1, 2015
|
|
7XRR
 
 | | Crystal structure of the human OX2R bound to the insomnia drug lemborexant. | | Descriptor: | (1~{R},2~{S})-2-[(2,4-dimethylpyrimidin-5-yl)oxymethyl]-~{N}-(5-fluoranylpyridin-2-yl)-2-(3-fluorophenyl)cyclopropane-1-carboxamide, Orexin receptor type 2 | | Authors: | Asada, H, Im, D, Iwata, S. | | Deposit date: | 2022-05-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Molecular basis for anti-insomnia drug design from structure of lemborexant-bound orexin 2 receptor.
Structure, 30, 2022
|
|
5ODQ
 
 | | Heterodisulfide reductase / [NiFe]-hydrogenase complex from Methanothermococcus thermolithotrophicus soaked with bromoethanesulfonate. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-bromanylethanesulfonic acid, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Wagner, T, Koch, J, Ermler, U, Shima, S. | | Deposit date: | 2017-07-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Methanogenic heterodisulfide reductase (HdrABC-MvhAGD) uses two noncubane [4Fe-4S] clusters for reduction.
Science, 357, 2017
|
|
