1S38
 
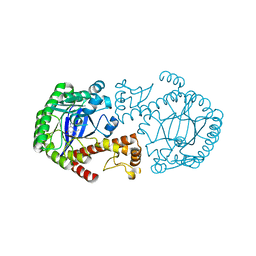 | | CRYSTAL STRUCTURE OF TGT IN COMPLEX WITH 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE | | Descriptor: | 2-AMINO-8-METHYLQUINAZOLIN-4(3H)-ONE, ZINC ION, tRNA guanine transglycosylase | | Authors: | Meyer, E.A, Furler, M, Diederich, F, Brenk, R, Klebe, G. | | Deposit date: | 2004-01-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis and In vitro Evaluation of 2-Aminoquinazolin-4(3H)-one-based Inhibitors for tRNA-Guanine Transglycosylase (TGT)
HELV.CHIM.ACTA, 87, 2004
|
|
3I2X
 
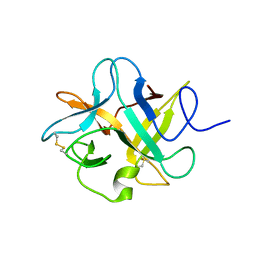 | | Crystal structure of a chimeric trypsin inhibitor having reactive site loop of ETI on the scaffold of WCI | | Descriptor: | Chymotrypsin inhibitor 3 | | Authors: | Sen, U, Khamrui, S, Dasgupta, J, Dattagupta, J.K, Majumder, S. | | Deposit date: | 2009-06-30 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Identification of a novel set of scaffolding residues that are instrumental for the inhibitory property of Kunitz (STI) inhibitors.
Protein Sci., 19, 2010
|
|
2Z7K
 
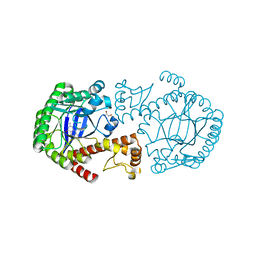 | | tRNA-Guanine transglycosylase (TGT) in complex with 2-Amino-lin-Benzoguanine | | Descriptor: | 2,6-diamino-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2007-08-24 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
3C2Y
 
 | | tRNA-Guanine Transglycosylase (TGT) in complex with 6-Amino-2-methyl-1,7-dihydro-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-methyl-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-01-26 | | Release date: | 2009-02-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure analysis and in silico pKa calculations suggest strong pKa shifts of ligands as driving force for high-affinity binding to TGT
Chembiochem, 10, 2009
|
|
3GE7
 
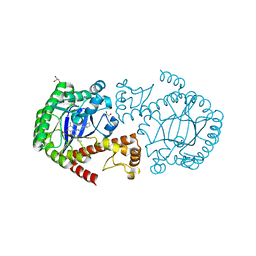 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclopentylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
3HFY
 
 | | Mutant of tRNA-guanine transglycosylase (K52M) | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ritschel, T, Klebe, G. | | Deposit date: | 2009-05-13 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Integrative Approach Combining Noncovalent Mass Spectrometry, Enzyme Kinetics and X-ray Crystallography to Decipher Tgt Protein-Protein and Protein-RNA Interaction
J.Mol.Biol., 393, 2009
|
|
2K95
 
 | | Solution structure of the wild-type P2B-P3 pseudoknot of human telomerase RNA | | Descriptor: | Telomerase RNA P2b-P3 pseudoknot | | Authors: | Kim, N.-K, Zhang, Q, Zhou, J, Theimer, C.A, Peterson, R.D, Feigon, J. | | Deposit date: | 2008-09-29 | | Release date: | 2008-11-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Dynamics of the Wild-type Pseudoknot of Human Telomerase RNA.
J.Mol.Biol., 384, 2008
|
|
3CI2
 
 | |
3EOU
 
 | | tRNA-guanine transglycosylase in complex with 6-amino-4-(2-hydroxyethyl)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-(2-hydroxyethyl)-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
3GC5
 
 | | tRNA-guanine transglycosylase in complex with 6-amino-4-(2-aminoethyl)-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-(2-aminoethyl)-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-21 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
3GC4
 
 | | tRNA-guanine transglycosylase in complex with inhibitor | | Descriptor: | 6-amino-4-[2-(benzylamino)ethyl]-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2009-02-21 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
3EOS
 
 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
1SVP
 
 | | SINDBIS VIRUS CAPSID PROTEIN | | Descriptor: | SINDBIS VIRUS CAPSID PROTEIN | | Authors: | Lee, S, Rossmann, M.G. | | Deposit date: | 1996-03-22 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly.
Structure, 4, 1996
|
|
4K1S
 
 | | Gly-Ser-SplB protease from Staphylococcus aureus at 1.96 A resolution | | Descriptor: | Serine protease SplB | | Authors: | Zdzalik, M, Pustelny, K, Stec-Niemczyk, J, Cichon, P, Czarna, A, Popowicz, G, Drag, M, Wladyka, B, Potempa, J, Dubin, A, Dubin, G. | | Deposit date: | 2013-04-05 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Staphylococcal SplB Serine Protease Utilizes a Novel Molecular Mechanism of Activation.
J.Biol.Chem., 289, 2014
|
|
8B49
 
 | | STRUCTURE OF PORCINE PANCREATIC ELASTASE BOUND TO A FRAGMENT (m-toluoylcarbonyl group) OF A 5-AZAINDOLE INHIBITOR | | Descriptor: | 1-(3-methylphenyl)carbonylpyrrolo[3,2-c]pyridine-3-carbonitrile, CALCIUM ION, Chymotrypsin-like elastase family member 1, ... | | Authors: | Ferraroni, M, Giovannoni, P, Gerace, A. | | Deposit date: | 2022-09-20 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | X-ray structural study of human neutrophil elastase inhibition with a series of azaindoles, azaindazoles and isoxazolones
J.Mol.Struct., 1274, 2023
|
|
1EXF
 
 | | EXFOLIATIVE TOXIN A | | Descriptor: | EXFOLIATVE TOXIN A, GLYCINE | | Authors: | Vath, G.M, Earhart, C.A, Rago, J.V, Kim, M.H, Bohach, G.A, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-10-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the superantigen exfoliative toxin A suggests a novel regulation as a serine protease.
Biochemistry, 36, 1997
|
|
8B53
 
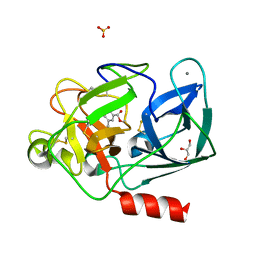 | |
1ELA
 
 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-prolinamide, ACETIC ACID, CALCIUM ION, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-04-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1QNJ
 
 | | THE STRUCTURE OF NATIVE PORCINE PANCREATIC ELASTASE AT ATOMIC RESOLUTION (1.1 A) | | Descriptor: | ELASTASE, SODIUM ION, SULFATE ION | | Authors: | Wurtele, M, Hahn, M, Hilpert, K, Hohne, W. | | Deposit date: | 1999-10-15 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Native Porcine Pancreatic Elastase at 1.1 A
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1ELE
 
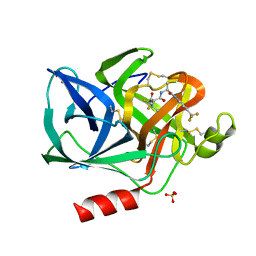 | | STRUCTURAL ANALYSIS OF THE ACTIVE SITE OF PORCINE PANCREATIC ELASTASE BASED ON THE X-RAY CRYSTAL STRUCTURES OF COMPLEXES WITH TRIFLUOROACETYL-DIPEPTIDE-ANILIDE INHIBITORS | | Descriptor: | CALCIUM ION, ELASTASE, N-(trifluoroacetyl)-L-valyl-N-[4-(trifluoromethyl)phenyl]-L-alaninamide, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
1ELB
 
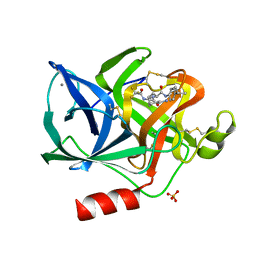 | | Analogous inhibitors of elastase do not always bind analogously | | Descriptor: | 6-ammonio-N-(trifluoroacetyl)-L-norleucyl-N-[4-(1-methylethyl)phenyl]-L-leucinamide, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Rasmussen, B, Ding, X, Petsko, G.A, Ringe, D. | | Deposit date: | 1993-12-07 | | Release date: | 1994-06-22 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analogous inhibitors of elastase do not always bind analogously.
Nat.Struct.Biol., 1, 1994
|
|
1ELD
 
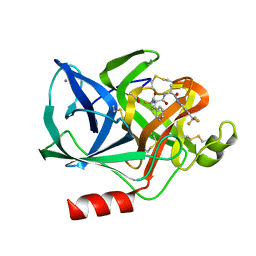 | | Structural analysis of the active site of porcine pancreatic elastase based on the x-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors | | Descriptor: | ACETIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Mattos, C, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-10-24 | | Release date: | 1995-02-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the active site of porcine pancreatic elastase based on the X-ray crystal structures of complexes with trifluoroacetyl-dipeptide-anilide inhibitors.
Biochemistry, 34, 1995
|
|
7P6T
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form III) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethyl-2-(hydroxymethyl)propane-1,3-diol, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6S
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2, pentane-1,5-diol | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7P6R
 
 | | Crystal structure of the FimH-binding decoy module of human glycoprotein 2 (GP2) (crystal form I) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform Alpha of Pancreatic secretory granule membrane major glycoprotein GP2 | | Authors: | Stsiapanava, A, Tunyasuvunakool, K, Jumper, J, de Sanctis, D, Jovine, L. | | Deposit date: | 2021-07-17 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the decoy module of human glycoprotein 2 and uromodulin and its interaction with bacterial adhesin FimH.
Nat.Struct.Mol.Biol., 29, 2022
|
|
