6IRW
 
 | | Crystal structure of the human cap-specific adenosine methyltransferase bound to SAH | | Descriptor: | Phosphorylated CTD-interacting factor 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-14 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cap-specific terminal N 6 -methylation of RNA by an RNA polymerase II-associated methyltransferase.
Science, 363, 2019
|
|
3ER9
 
 | | Crystal structure of the heterodimeric vaccinia virus mRNA polyadenylate polymerase complex with UU and 3'-deoxy ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, 5'-R(UP*U)-3', CALCIUM ION, ... | | Authors: | Li, C, Li, H, Zhou, S, Poulos, T.L, Gershon, P.D. | | Deposit date: | 2008-10-01 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Polymerase Translocation with Respect to Single-Stranded Nucleic Acid: Looping or Wrapping of Primer around a Poly(A) Polymerase
Structure, 17, 2009
|
|
3MF5
 
 | |
7UMR
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with compound SJ001034732-1 (trans-form) | | Descriptor: | (1P,18Z)-5-hydroxy-16,21-dioxa-3,8,28-triazatetracyclo[20.3.1.1~2,6~.1~11,15~]octacosa-1(26),2(28),5,11(27),12,14,18,22,24-nonaene-4,7-dione, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2022-04-07 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
2O1P
 
 | |
6P1S
 
 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated AMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
7EGQ
 
 | | Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L.M, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading.
Cell, 184, 2021
|
|
6P1U
 
 | | Post-catalytic nicked complex of human DNA Polymerase Mu with 1-nt gapped substrate containing template 8OG and newly incorporated CMP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kaminski, A.M, Pedersen, L.C, Bebenek, K, Chiruvella, K.K, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2019-05-20 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Unexpected behavior of DNA polymerase Mu opposite template 8-oxo-7,8-dihydro-2'-guanosine.
Nucleic Acids Res., 47, 2019
|
|
7ZPY
 
 | |
8JLA
 
 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
6C6S
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6U
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with NusG | | Descriptor: | DNA (29-MER), DNA-DIRECTED RNA POLYMERASE BETA', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
6C6T
 
 | | CryoEM structure of E.coli RNA polymerase elongation complex bound with RfaH | | Descriptor: | DNA (29-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kang, J.Y, Artsimovitch, I, Landick, R, Darst, S.A. | | Deposit date: | 2018-01-19 | | Release date: | 2018-07-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators.
Cell, 173, 2018
|
|
4KBM
 
 | |
5ZIT
 
 | | Crystal structure of human Enterovirus D68 RdRp in complex with NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, RdRp | | Authors: | Wang, M.L, Li, L, Chen, Y.P, Jiang, H, Zhang, Y, Su, D. | | Deposit date: | 2018-03-17 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Structure of the enterovirus D68 RNA-dependent RNA polymerase in complex with NADPH implicates an inhibitor binding site in the RNA template tunnel.
J.Struct.Biol., 2020
|
|
4NZ0
 
 | |
4NYZ
 
 | | The EMCV 3Dpol structure with altered motif A conformation at 2.15A resolution | | Descriptor: | GLUTAMINE, GLYCEROL, Genome polyprotein, ... | | Authors: | Vives-adrian, L, Lujan, C, Ferrer-orta, C, Verdaguer, N. | | Deposit date: | 2013-12-11 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a cardiovirus RNA-dependent RNA polymerase reveals an unusual conformation of the polymerase active site
J.Virol., 88, 2014
|
|
7EIZ
 
 | | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 10, ... | | Authors: | Yan, L, Yang, Y.X, Li, M.Y, Zhang, Y, Zheng, L.T, Ge, J, Huang, Y.C, Liu, Z.Y, Wang, T, Gao, S, Zhang, R, Huang, Y.Y, Guddat, L.W, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2021-04-01 | | Release date: | 2021-09-22 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Cell, 184, 2021
|
|
2BRK
 
 | |
2BRL
 
 | |
6ASG
 
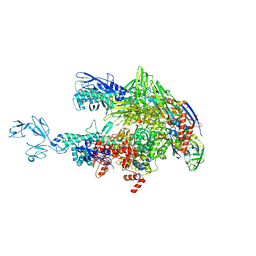 | | Crystal structure of Thermus thermophilus RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, Y, Lin, W, Ying, R, Ebright, R.H. | | Deposit date: | 2017-08-24 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Basis of Transcription Inhibition by Fidaxomicin (Lipiarmycin A3).
Mol. Cell, 70, 2018
|
|
7X6S
 
 | |
4YD2
 
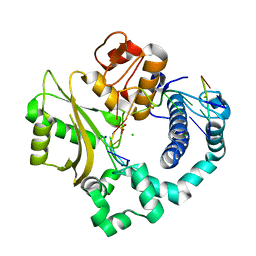 | | Nicked complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*CP*GP*TP*AP*T)-3'), ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.471 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4YCX
 
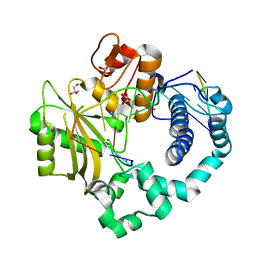 | | Binary complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*AP*TP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*A)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4LQ9
 
 | | Crystal structure of human norovirus RNA-dependent RNA-polymerase in complex with NAF2 | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA-polymerase, naphthalene-1,5-disulfonic acid | | Authors: | Milani, M, Tarantino, D, Mastrangelo, E, Croci, R. | | Deposit date: | 2013-07-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naphthalene-sulfonate inhibitors of human norovirus RNA-dependent RNA-polymerase.
Antiviral Res., 102, 2014
|
|
