4FLS
 
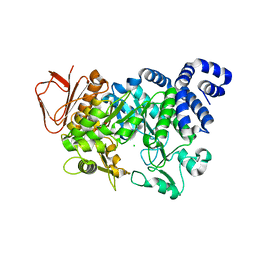 | | Crystal structure of Amylosucrase inactive double mutant F290K-E328Q from Neisseria polysaccharea in complex with sucrose. | | Descriptor: | Amylosucrase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Guerin, F, Champion, E, Moulis, C, Barbe, S, Tran, T.H, Morel, S, Descroix, K, Monsan, P, Mulard, L.A, Remaud-Simeon, M, Andre, I, Mourey, L, Tranier, S. | | Deposit date: | 2012-06-15 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Applying pairwise combinations of amino Acid mutations for sorting out highly efficient glucosylation tools for chemo-enzymatic synthesis of bacterial oligosaccharides.
J.Am.Chem.Soc., 134, 2012
|
|
3CI2
 
 | |
4FNU
 
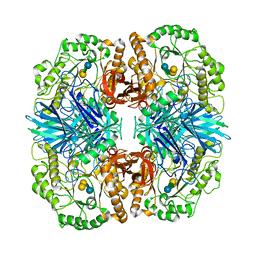 | | Crystal structure of GH36 alpha-galactosidase AgaA A355E D478A from Geobacillus stearothermophilus in complex with stachyose | | Descriptor: | Alpha-galactosidase AgaA, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose-(1-6)-alpha-D-galactopyranose-(1-6)-alpha-D-galactopyranose | | Authors: | Merceron, R, Foucault, M, Haser, R, Mattes, R, Watzlawick, H, Gouet, P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The molecular mechanism of the thermostable alpha-galactosidases AgaA and AgaB explained by X-ray crystallography and mutational studies
J.Biol.Chem., 287, 2012
|
|
3CG7
 
 | |
4FM4
 
 | | Wild Type Fe-type Nitrile Hydratase from Comamonas testosteroni Ni1 | | Descriptor: | FE (III) ION, Nitrile hydratase alpha subunit, Nitrile hydratase beta subunit, ... | | Authors: | Kuhn, M.L, Martinez, S, Gumataotao, N, Bornscheuer, U, Liu, D, Holz, R.C. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | The Fe-type nitrile hydratase from Comamonas testosteroni Ni1 does not require an activator accessory protein for expression in Escherichia coli.
Biochem.Biophys.Res.Commun., 424, 2012
|
|
4FOJ
 
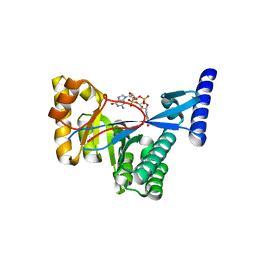 | | 1.55 A Crystal Structure of Xanthomonas citri FimX EAL domain in complex with c-diGMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), FimX | | Authors: | Farah, C.S, Guzzo, C.R. | | Deposit date: | 2012-06-20 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the PilZ-FimXEAL-c-di-GMP Complex Responsible for the Regulation of Bacterial Type IV Pilus Biogenesis.
J.Mol.Biol., 425, 2013
|
|
3CGO
 
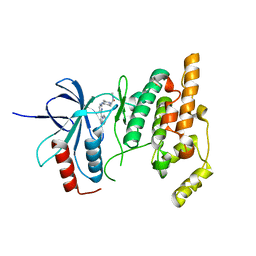 | | IRAK-4 Inhibitors (Part II)- A structure based assessment of imidazo[1,2 a]pyridine binding | | Descriptor: | 2-{4-[(4-imidazo[1,2-a]pyridin-3-ylpyrimidin-2-yl)amino]piperidin-1-yl}-N-methylacetamide, Mitogen-activated protein kinase 10 | | Authors: | Ceska, T.A, Platt, A, Fortunato, M, Dickson, K.M, Beevers, R. | | Deposit date: | 2008-03-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | IRAK-4 inhibitors. Part II: A structure-based assessment of imidazo[1,2-a]pyridine binding
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CGT
 
 | |
4FNJ
 
 | |
4FNN
 
 | | Crystal structure of the complex of CPGRP-S with stearic acid at 2.2 A RESOLUTION | | Descriptor: | Peptidoglycan recognition protein 1, STEARIC ACID | | Authors: | Dube, D, Sharma, P, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural basis of the binding of fatty acids to peptidoglycan recognition protein, PGRP-S through second binding site.
Arch.Biochem.Biophys., 529, 2013
|
|
3CHE
 
 | |
3CHH
 
 | | Crystal Structure of Di-iron AurF | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-09 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CHQ
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with N5-[4-(phenylmethoxy)phenyl]-L-glutamine | | Descriptor: | (2S)-2-amino-5-oxo-5-[(4-phenylmethoxyphenyl)amino]pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
6QQO
 
 | | Aplysia californica AChBP in complex with 2-Fluoro-(carbamoylpyridinyl)deschloroepibatidine analogue (3) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[5-[(1~{R},2~{R},4~{S})-7-azabicyclo[2.2.1]heptan-2-yl]-2-fluoranyl-pyridin-3-yl]pyridine-3-carboxamide, ... | | Authors: | Bueno, R.V, Davis, S, Dawson, A, Hunter, W.N. | | Deposit date: | 2019-02-18 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Interactions between 2'-fluoro-(carbamoylpyridinyl)deschloroepibatidine analogues and acetylcholine-binding protein inform on potent antagonist activity against nicotinic receptors
Acta Crystallogr.,Sect.D, 78, 2022
|
|
4FO2
 
 | | Heat-labile enterotoxin LT-IIb-B5(T13I) mutant | | Descriptor: | ACETATE ION, Heat-labile enterotoxin IIB, B chain, ... | | Authors: | Cody, V, Pace, J, Nawar, H, Liang, S, Connell, T, Hajishengallis, G. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-activity correlations of variant forms of the B pentamer of Escherichia coli type II heat-labile enterotoxin LT-IIb with Toll-like receptor 2 binding.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3CIW
 
 | | X-RAY structure of the [FeFe]-hydrogenase maturase HydE from thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FeFe-Hydrogenase maturase, ... | | Authors: | Nicolet, Y, Ruback, J.K, Posewitz, M.C, Amara, P, Mathevon, C, Atta, M, Fontecave, M, Fontecilla-Camps, J.C. | | Deposit date: | 2008-03-12 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | X-ray Structure of the [FeFe]-Hydrogenase Maturase HydE from Thermotoga maritima
J.Biol.Chem., 283, 2008
|
|
4FOP
 
 | | Crystal Structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 1.86 A resolution | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kaushik, S, Kumar, S, Singh, N, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-06-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
3CET
 
 | | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63 | | Descriptor: | Conserved archaeal protein, SULFATE ION | | Authors: | Kuzin, A.P, Su, M, Seetharaman, J, Forouhar, F, Wang, D, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, J.F, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the pantheonate kinase-like protein Q6M145 at the resolution 1.8 A. Northeast Structural Genomics Consortium target MrR63.
To be Published
|
|
3CJ9
 
 | |
3CJF
 
 | | Crystal structure of VEGFR2 in complex with a 3,4,5-trimethoxy aniline containing pyrimidine | | Descriptor: | N~4~-(3-methyl-1H-indazol-6-yl)-N~2~-(3,4,5-trimethoxyphenyl)pyrimidine-2,4-diamine, SULFATE ION, Vascular endothelial growth factor receptor 2 | | Authors: | Nolte, R.T. | | Deposit date: | 2008-03-12 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 5-[[4-[(2,3-dimethyl-2H-indazol-6-yl)methylamino]-2-pyrimidinyl]amino]-2-methyl-benzenesulfonamide (Pazopanib), a novel and potent vascular endothelial growth factor receptor inhibitor.
J.Med.Chem., 51, 2008
|
|
4FD4
 
 | |
3VYF
 
 | | Human renin in complex with inhibitor 9 | | Descriptor: | (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]-N-(2,6-dimethylheptan-4-yl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3CJT
 
 | | Ribosomal protein L11 methyltransferase (PrmA) in complex with dimethylated ribosomal protein L11 | | Descriptor: | 1,2-ETHANEDIOL, 50S ribosomal protein L11, CHLORIDE ION, ... | | Authors: | Demirci, H, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2008-03-13 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple-Site Trimethylation of Ribosomal Protein L11 by the PrmA Methyltransferase.
Structure, 16, 2008
|
|
6OJQ
 
 | |
3CFJ
 
 | |
