8U4P
 
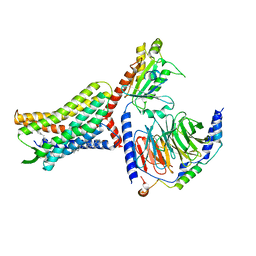 | | Structure of AMD3100-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4O
 
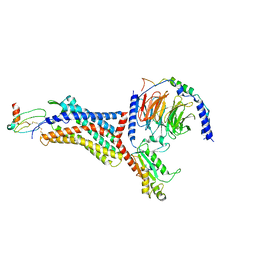 | | Structure of CXCL12-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
6ENT
 
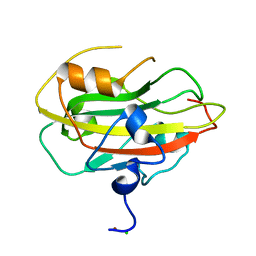 | | Structure of the rat RKIP variant delta143-146 | | Descriptor: | CHLORIDE ION, Phosphatidylethanolamine-binding protein 1, ZINC ION | | Authors: | Koelmel, W, Hirschbeck, M, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8UEV
 
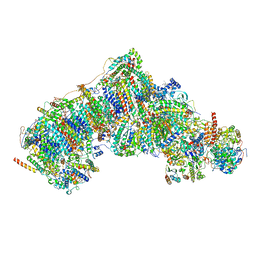 | | In-situ complex I, Deactive class04 | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Zheng, W, Zhu, J, Zhang, K. | | Deposit date: | 2023-10-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
6L87
 
 | |
6S3W
 
 | | Solution NMR Structure of TolAIII Bound to a Peptide Derived from the N-terminus of TolB | | Descriptor: | Cell envelope integrity/translocation protein TolA, TolBp | | Authors: | Kleanthous, C, Redfield, C, Rajasekar, K, Holmes, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-03-25 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | The lipoprotein Pal stabilises the bacterial outer membrane during constriction by a mobilisation-and-capture mechanism.
Nat Commun, 11, 2020
|
|
8UEE
 
 | | Atomic structure of Salmonella SipA/F-actin complex by cryo-EM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Niedzialkowska, E, Runyan, L, Kudryashova, E, Kudryashov, D.S, Egelman, E.H. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stabilization of F-actin by Salmonella effector SipA resembles the structural effects of inorganic phosphate and phalloidin.
Structure, 32, 2024
|
|
7MZZ
 
 | |
2F35
 
 | |
6PIG
 
 | | V. cholerae TniQ-Cascade complex, closed conformation | | Descriptor: | RNA (60-MER), TniQ monomer 1, TniQ monomer 2, ... | | Authors: | Halpin-Healy, T, Klompe, S, Sternberg, S.H. | | Deposit date: | 2019-06-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Nature, 577, 2020
|
|
4LZO
 
 | | Crystal structure of human PRS1 A87T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
5WUA
 
 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
4LYG
 
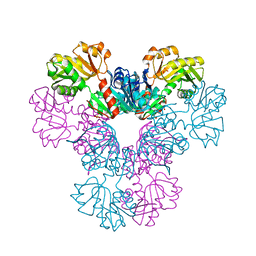 | | Crystal structure of human PRS1 E43T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
6S7H
 
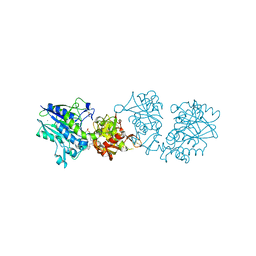 | | Human CD73 (5'-nucleotidase) in complex with PSB12489 (an AOPCP derivative) in the closed state | | Descriptor: | (N6,N6)-methyl,benzyl-C2-chloro-(alpha,beta)-methylene-ADP, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-Ray Co-Crystal Structure Guides the Way to Subnanomolar Competitive Ecto-5'-Nucleotidase (CD73) Inhibitors for Cancer Immunotherapy
Adv Ther, 2019
|
|
4LZN
 
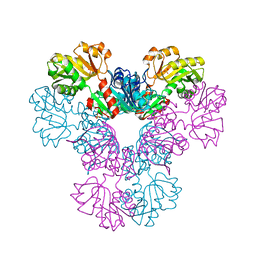 | | Crystal structure of human PRS1 D65N mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-07-31 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
4M0P
 
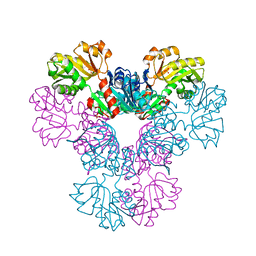 | | Crystal structure of human PRS1 M115T mutant | | Descriptor: | Ribose-phosphate pyrophosphokinase 1, SULFATE ION | | Authors: | Chen, P, Teng, M, Li, X. | | Deposit date: | 2013-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal and EM Structures of Human Phosphoribosyl Pyrophosphate Synthase I (PRS1) Provide Novel Insights into the Disease-Associated Mutations
Plos One, 10, 2015
|
|
5EPQ
 
 | | Structure at 1.75 A resolution of a glycosylated, lipid-binding, lipocalin-like protein | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Banerjee, S, Chavas, L.M.G, Ramaswamy, S. | | Deposit date: | 2015-11-12 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structure of a heterogeneous, glycosylated, lipid-bound, in vivo-grown protein crystal at atomic resolution from the viviparous cockroach Diploptera punctata.
Iucrj, 3, 2016
|
|
6RM2
 
 | | Deoxyguanylosuccinate synthase (DgsS) structure with ATP, IMP, Magnesium | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Adenylosuccinate synthetase, INOSINIC ACID, ... | | Authors: | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with ATP, IMP, Magnesium at 2.5 Angstrom resolution
To Be Published
|
|
6S51
 
 | | The crystal structure of glycogen phosphorylase in complex with 10 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S4K
 
 | | The crystal structure of glycogen phosphorylase in complex with 12 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-phenyl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
2FZ8
 
 | | Human Aldose reductase complexed with inhibitor zopolrestat at 1.48 A(1 day soaking). | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
6S4O
 
 | | The crystal structure of glycogen phosphorylase in complex with 9 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-naphthalen-2-yl-1~{H}-imidazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
4MH8
 
 | |
6RY9
 
 | |
2FZB
 
 | | Human Aldose Reductase complexed with four tolrestat molecules at 1.5 A resolution. | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TOLRESTAT, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
