1T7R
 
 | | Crystal structure of the androgen receptor ligand binding domain in complex with a FxxLF motif | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, FxxLF motif peptide | | Authors: | Hur, E, Pfaff, S.J, Payne, E.S, Gron, H, Buehrer, B.M, Fletterick, R.J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition and accommodation at the androgen receptor coactivator binding interface.
Plos Biol., 2, 2004
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
7DP5
 
 | | Crystal structure of mutant V45A Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate and methotrexate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
6RKM
 
 | | Fragment AZ-022 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-phenylazanyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
7DP6
 
 | | Crystal structure of mutant V45T Brugia malayi thymidylate synthase complexed with 2'-deoxyuridine monophosphate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Keyunratsami, K, Nualnoi, T, Wongkamchai, S, Songsiriritthigul, C, Chen, C.-J, Canyuk, B. | | Deposit date: | 2020-12-18 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic investigation and inhibition of Brugia malayi thymidylate synthase by a folate analog
To Be Published
|
|
2LLP
 
 | | Solution structure of a THP type 1 alpha 1 collagen fragment (772-786) | | Descriptor: | Collagen alpha-1(I) chain | | Authors: | Bertini, I, Fragai, M, Luchinat, C, Melikian, M, Toccafondi, M, Lauer, J.L, Fields, G.B, Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for matrix metalloproteinase 1-catalyzed collagenolysis.
J.Am.Chem.Soc., 134, 2012
|
|
7JYX
 
 | | Crystal Structure of HLA A*2402 in complex with TYQWIIRNWET, an 11-mer epitope from Influenza | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, PB2 peptide from Influenza, ... | | Authors: | Gras, S, Nguyen, A.T, Szeto, C, Rossjohn, J. | | Deposit date: | 2020-09-01 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CD8 + T cell landscape in Indigenous and non-Indigenous people restricted by influenza mortality-associated HLA-A*24:02 allomorph.
Nat Commun, 12, 2021
|
|
4IYA
 
 | | Structure of the Y250A mutant of the PANTON-VALENTINE LEUCOCIDIN S component from STAPHYLOCOCCUS AUREUS | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, LukS-PV | | Authors: | Maveyraud, L, Guerin, F, Laventie, B.J, Prevost, G, Mourey, L. | | Deposit date: | 2013-01-28 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Residues essential for panton-valentine leukocidin s component binding to its cell receptor suggest both plasticity and adaptability in its interaction surface
Plos One, 9, 2014
|
|
6XLK
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-4nt | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
4IYL
 
 | | 30S ribosomal protein S15 from Campylobacter jejuni | | Descriptor: | 30S ribosomal protein S15 | | Authors: | Osipiuk, J, Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-28 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | 30S ribosomal protein S15 from Campylobacter jejuni
To be Published
|
|
6XLA
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
5MH4
 
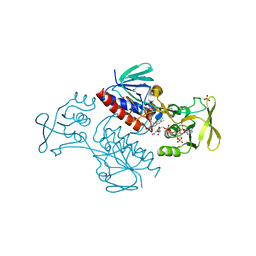 | | Crystal Structure of Lactococcus lactis Thioredoxin Reductase (FR conformation) | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Skjoldager, N, Bang, M.B, Svensson, B, Hagglund, P, Harris, P. | | Deposit date: | 2016-11-23 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The structure of Lactococcus lactis thioredoxin reductase reveals molecular features of photo-oxidative damage.
Sci Rep, 7, 2017
|
|
6XL6
 
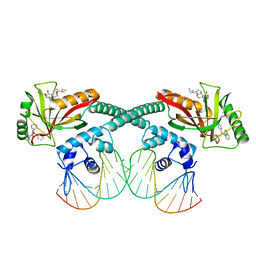 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPo | | Descriptor: | CHAPSO, MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, ... | | Authors: | Yang, Y, Liu, C, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
4RX3
 
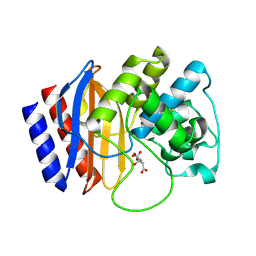 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | Descriptor: | Beta-lactamase TEM, CITRATE ANION | | Authors: | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2014-12-08 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4IZW
 
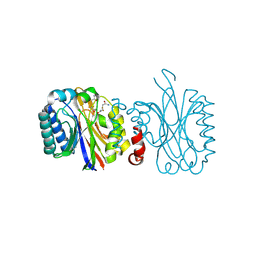 | |
6V83
 
 | | Crystal structure of CTX-M-14 E166A/P167S/D240G beta-lactamase in complex with ceftazidime-1 | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Brown, C.A, Hu, L, Sankaran, B, Prasad, B.V.V, Palzkill, T.G. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Antagonism between substitutions in beta-lactamase explains a path not taken in the evolution of bacterial drug resistance.
J.Biol.Chem., 295, 2020
|
|
6RSR
 
 | | TBK1 in complex with compound 2 | | Descriptor: | Serine/threonine-protein kinase TBK1, ~{N}-(cyclopropen-1-ylmethyl)-2-[[4-[[4-[3,3,3-tris(fluoranyl)propanoyl]piperazin-1-yl]methyl]pyridin-2-yl]amino]-1~{H}-benzimidazole-5-carboxamide | | Authors: | Panne, D, Hillig, R.C, Rengachari, S. | | Deposit date: | 2019-05-22 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Discovery of BAY-985, a Highly Selective TBK1/IKK epsilon Inhibitor.
J.Med.Chem., 63, 2020
|
|
6V8O
 
 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
6VCW
 
 | | Crystal structure of Medicago truncatula S-adenosylmethionine Synthase 3A (MtMAT3A) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sekula, B, Ruszkowski, M, Dauter, Z. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | S-adenosylmethionine synthases in plants: Structural characterization of type I and II isoenzymes from Arabidopsis thaliana and Medicago truncatula.
Int.J.Biol.Macromol., 151, 2020
|
|
5MIX
 
 | |
4J29
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR258. | | Descriptor: | Engineered Protein OR258 | | Authors: | Vorobiev, S, Su, M, Koga, R, Seetharaman, J, Koga, N, Mao, L, Xiao, R, Kohan, E, Castelllanos, J, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-04 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Engineered Protein OR258.
To be Published
|
|
4X4X
 
 | |
6RWF
 
 | |
3E9A
 
 | | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, SULFATE ION | | Authors: | Nocek, B, Mulligan, R, Kwon, K, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-08-21 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from Vibrio cholerae O1
biovar eltor str. N16961
To be Published
|
|
6RX2
 
 | | Fragment AZ-005 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-[[(2~{S})-1-azanylpropan-2-yl]amino]-1-benzothiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-07 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
