4AQR
 
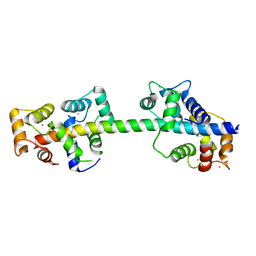 | | Crystal structure of calmodulin in complex with the regulatory domain of a plasma-membrane Ca2+-ATPase | | Descriptor: | CALCIUM ION, CALCIUM-TRANSPORTING ATPASE 8, PLASMA MEMBRANE-TYPE, ... | | Authors: | Tidow, H, Poulsen, L.R, Andreeva, A, Hein, K.L, Palmgren, M.G, Nissen, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Bimodular Mechanism of Calcium Control in Eukaryotes
Nature, 491, 2012
|
|
4YYK
 
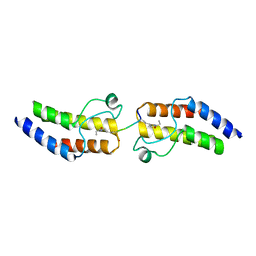 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6I7E
 
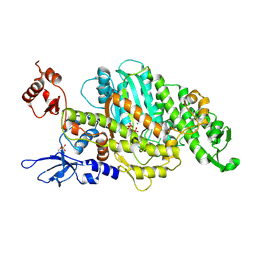 | | Plasmodium falciparum Myosin A, Pre-powerstroke | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin-A, ... | | Authors: | Robert-Paganin, J, Auguin, D, Moussaoui, D, Jousset, G, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2018-11-16 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.492 Å) | | Cite: | Plasmodium myosin A drives parasite invasion by an atypical force generating mechanism.
Nat Commun, 10, 2019
|
|
5X5I
 
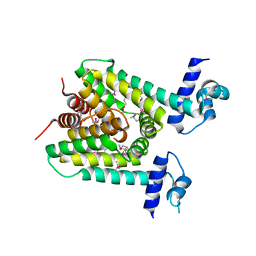 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
3KMZ
 
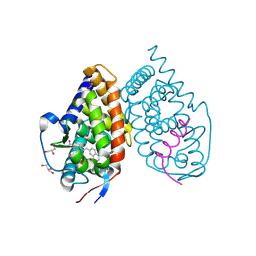 | | Crystal structure of RARalpha ligand binding domain in complex with the inverse agonist BMS493 and a corepressor fragment | | Descriptor: | 4-{(E)-2-[5,5-dimethyl-8-(phenylethynyl)-5,6-dihydronaphthalen-2-yl]ethenyl}benzoic acid, GLYCEROL, Nuclear receptor corepressor 1, ... | | Authors: | Bourguet, W, le Maire, A. | | Deposit date: | 2009-11-11 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A unique secondary-structure switch controls constitutive gene repression by retinoic acid receptor.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5W9H
 
 | |
3HP8
 
 | | Crystal structure of a designed Cyanovirin-N homolog lectin; LKAMG, bound to sucrose | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Cyanovirin-N-like protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Koharudin, L.M.I, Furey, W, Gronenborn, A.M. | | Deposit date: | 2009-06-03 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A designed chimeric cyanovirin-N homolog lectin: Structure and molecular basis of sucrose binding.
Proteins, 77, 2009
|
|
2WPX
 
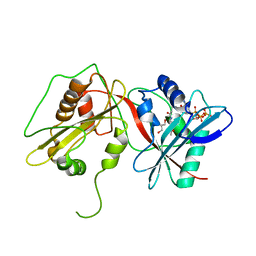 | | Tandem GNAT protein from the clavulanic acid biosynthesis pathway (with AcCoA) | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, ORF14 | | Authors: | Iqbal, A, Arunlanantham, H, McDonough, M.A, Chowdhury, R, Clifton, I.J. | | Deposit date: | 2009-08-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and mass spectrometric analyses of a tandem GNAT protein from the clavulanic acid biosynthesis pathway.
Proteins, 78, 2010
|
|
5ES1
 
 | |
2WV4
 
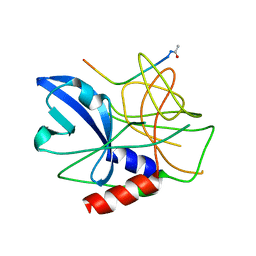 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
5G51
 
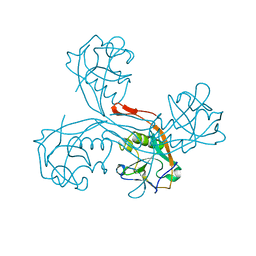 | | High resolution structure of the part of VP3 protein of Deformed Wing Virus forming P-domain | | Descriptor: | DWV-VP3-P-DOMAIN | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ZUZ
 
 | |
5W9I
 
 | |
4YYH
 
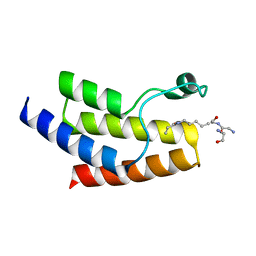 | | Crystal structure of BRD9 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
4YYN
 
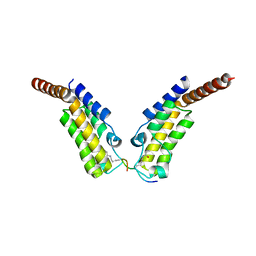 | | Crystal structure of TAF1 BD2 Bromodomain bound to a crotonyllysine peptide | | Descriptor: | Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
5W9L
 
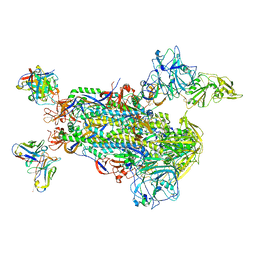 | |
6BZY
 
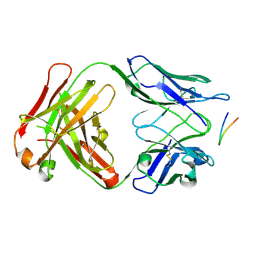 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4YYM
 
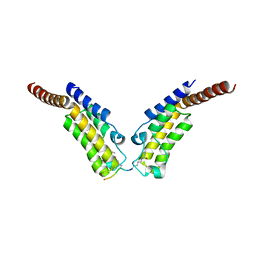 | | Crystal structure of TAF1 BD2 Bromodomain bound to a butyryllysine peptide | | Descriptor: | CALCIUM ION, Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
5ZX5
 
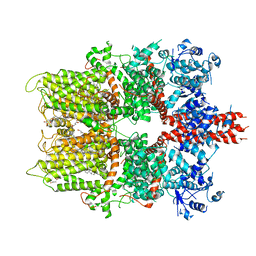 | | 3.3 angstrom structure of mouse TRPM7 with EDTA | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Transient receptor potential cation channel subfamily M member 7 | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | Deposit date: | 2018-05-18 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5W9K
 
 | |
4YYJ
 
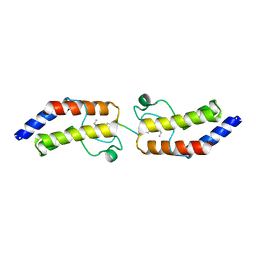 | | Crystal structure of BRD9 Bromodomain bound to a butyryllysine peptide | | Descriptor: | Bromodomain-containing protein 9, Histone H4 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-23 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
1Q78
 
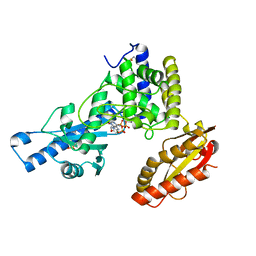 | | Crystal structure of poly(A) polymerase in complex with 3'-dATP and magnesium chloride | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Poly(A) polymerase alpha | | Authors: | Martin, G, Moglich, A, Keller, W, Doublie, S. | | Deposit date: | 2003-08-16 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase.
J.Mol.Biol., 341, 2004
|
|
5V7I
 
 | | Crystal structure of homo sapiens serine hydroxymethyltransferase 2 (mitochondrial) (SHMT2), in complex with glycine, PLP and folate-competitive pyrazolopyran inhibitor: 6-amino-4-isopropyl-3-methyl-4-(3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl)-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile | | Descriptor: | (4R)-6-amino-3-methyl-4-(propan-2-yl)-4-[3-(pyrrolidin-1-yl)-5-(trifluoromethyl)phenyl]-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ducker, G.S, Ghergurovich, J.M, Mainolfi, N, Suri, V, Jeong, S, Friedman, A, Manfredi, M, Kim, H, Rabinowitz, J.D. | | Deposit date: | 2017-03-20 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3HOP
 
 | | Structure of the actin-binding domain of human filamin A | | Descriptor: | Filamin-A, PHOSPHATE ION | | Authors: | Clark, A.R, Sawyer, G.M, Robertson, S.P, Sutherland-Smith, A.J. | | Deposit date: | 2009-06-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Skeletal dysplasias due to filamin A mutations result from a gain-of-function mechanism distinct from allelic neurological disorders
Hum.Mol.Genet., 18, 2009
|
|
1QFU
 
 | | INFLUENZA VIRUS HEMAGGLUTININ COMPLEXED WITH A NEUTRALIZING ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (HEMAGGLUTININ (HA1 CHAIN)), ... | | Authors: | Fleury, D, Gigant, B, Bizebard, T, Daniels, R.S, Skehel, J.J, Knossow, M. | | Deposit date: | 1999-04-14 | | Release date: | 1999-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A complex of influenza hemagglutinin with a neutralizing antibody that binds outside the virus receptor binding site.
Nat.Struct.Biol., 6, 1999
|
|
