8T4M
 
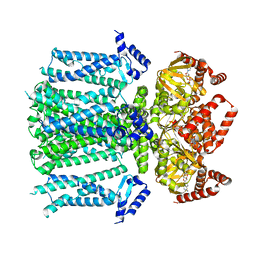 | | Closed human HCN1 F186C S264C bound to cAMP, reconstituted in LMNG + SPL | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1 | | Authors: | Burtscher, V, Mount, J, Cowgill, J, Chang, Y, Bickel, K, Yuan, P, Chanda, B. | | Deposit date: | 2023-06-09 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural basis for hyperpolarization-dependent opening of human HCN1 channel.
Nat Commun, 15, 2024
|
|
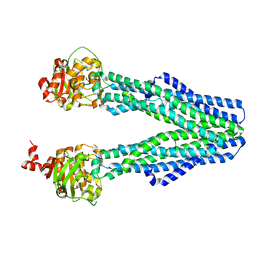
8T4J
 
 | |
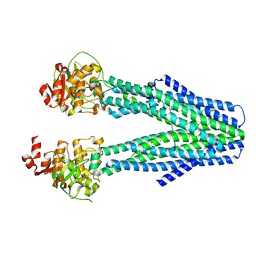
8T4G
 
 | |
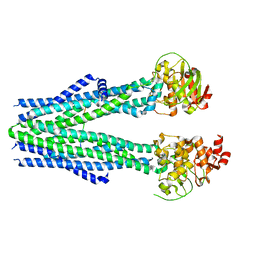
8T4F
 
 | |
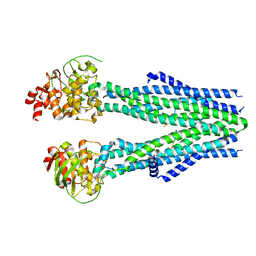
8T4I
 
 | |
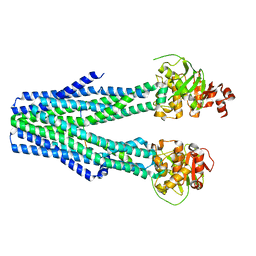
8T4H
 
 | |
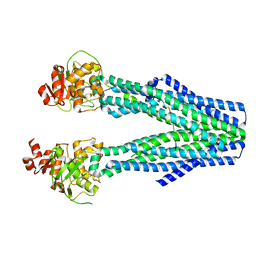
8T4E
 
 | |
8JOO
 
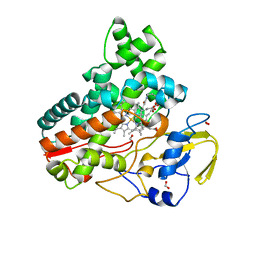 | | Crystal structure of cytochrome P450 IkaD from Streptomyces sp. ZJ306, in complex with the substrate ikarugamycin | | Descriptor: | (1Z,3E,5S,7R,8R,10R,11R,12S,15R,16S,18Z,25S)-11-ethyl-2-hydroxy-10-methyl-21,26-diazapentacyclo[23.2.1.05,16.07,15.08,12]octacosa-1(2),3,13,18-tetraene-20,27,28-trione, Cytochrome P450, FORMIC ACID, ... | | Authors: | Zhang, Y.L, Zhang, L.P, Zhang, C.S. | | Deposit date: | 2023-06-08 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Mechanistic Understanding of the Distinct Regio- and Chemoselectivity of Multifunctional P450s by Structural Comparison of IkaD and CftA Complexed with Common Substrates.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8T46
 
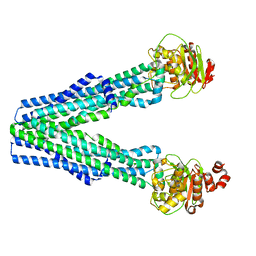 | |
8T3Y
 
 | | Structure of Bre1-nucleosome complex - state1 | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8PB1
 
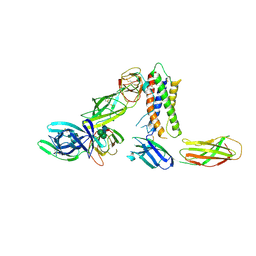 | | Cryo-EM structure of a pre-dimerized murine IL-12 complete extracellular signaling complex (Class 1), obtained after local refinement. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1,Death-associated protein kinase 1, Interleukin-12 receptor subunit beta-2,Calmodulin-1, ... | | Authors: | Felix, J, Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-06-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8T42
 
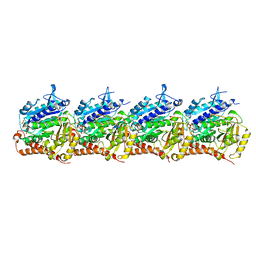 | | Model of TTLL6 MTBH1-2 bound to microtubule | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Mahalingan, K.K, Grotjahn, D, Li, Y, Lander, G.C, Zehr, E.A, Roll-Mecak, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for alpha-tubulin-specific and modification state-dependent glutamylation.
Nat.Chem.Biol., 2024
|
|
8PAU
 
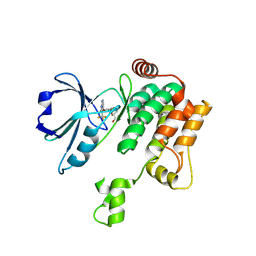 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, [(5~{R})-2-[[3,5-bis(fluoranyl)-4-[[3-(trifluoromethyl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]amino]-5-fluoranyl-4,6-dihydro-1,3-oxazin-5-yl]methanol | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAV
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1,3-thiazol-5-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, GLYCEROL, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAS
 
 | | Crystal structure of MAP4K1 with a SMOL inhibitor | | Descriptor: | 4-[2,6-bis(fluoranyl)-4-(3-morpholin-4-ylpropylcarbamoylamino)phenoxy]-~{N}-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]-1~{H}-pyrrolo[2,3-b]pyridine-3-carboxamide, Mitogen-activated protein kinase kinase kinase kinase 1 | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAW
 
 | | Crystal structure of MST1 with a MAP4K1 SMOL inhibitor | | Descriptor: | 1-[3,5-bis(fluoranyl)-4-[[3-(1-propan-2-ylpyrazol-3-yl)-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-3-(2-methoxyethyl)urea, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ASPARTIC ACID, ... | | Authors: | Friberg, A. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8PAR
 
 | | Crystal structure of human MAP4K1 with an inhibitor, BAY-405 | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase kinase kinase kinase 1, ~{N}-[3,5-bis(fluoranyl)-4-[[3-[1-(trifluoromethyl)cyclopropyl]-1~{H}-pyrrolo[2,3-b]pyridin-4-yl]oxy]phenyl]-2,9-dioxa-4-azaspiro[5.5]undec-3-en-3-amine | | Authors: | Schaefer, M. | | Deposit date: | 2023-06-08 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and optimization of Azaindole based MAP4K1 Inhibitors and the discovery of BAY-405
To Be Published
|
|
8JOH
 
 | |
8JO4
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8JO3
 
 | | Cryo-EM structure of a Legionella effector complexed with actin and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Zhou, X.T, Wang, X.F, Tan, J.X, Zhu, Y.Q. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Legionella effector LnaB is a phosphoryl AMPylase that impairs phosphosignalling.
Nature, 631, 2024
|
|
8T3V
 
 | | Cryo-EM structure of the DHA bound FFA1-Gq complex | | Descriptor: | CHOLESTEROL, DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 1, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
8T3W
 
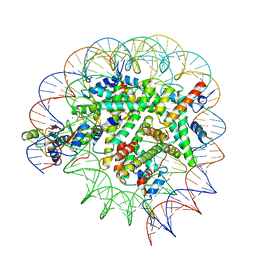 | | Structure of Bre1-nucleosome complex - state2 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3T
 
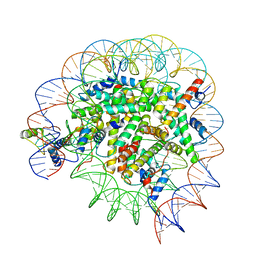 | | Structure of Bre1-nucleosome complex - state3 | | Descriptor: | 601 DNA strand 1, 601 DNA strand 2, E3 ubiquitin-protein ligase BRE1, ... | | Authors: | Zhao, F, Hicks, C.W, Wolberger, C. | | Deposit date: | 2023-06-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Mechanism of histone H2B monoubiquitination by Bre1.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8T3K
 
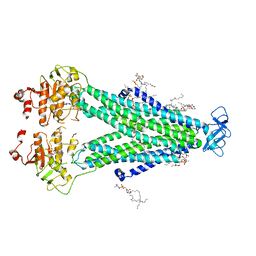 | |
8T3Q
 
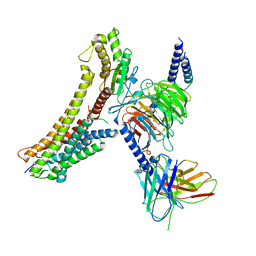 | | Cryo-EM structure of the DHA bound FFA4-Gq complex | | Descriptor: | DOCOSA-4,7,10,13,16,19-HEXAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
