7SLY
 
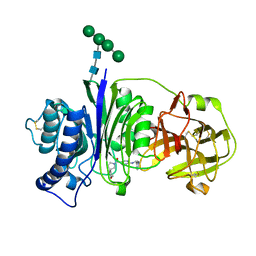 | | Vanin-1 complexed with Compound 27 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(1S)-1-(pyrazin-2-yl)ethyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLX
 
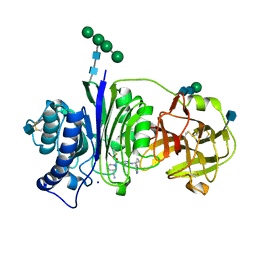 | | Vanin-1 complexed with Compound 11 | | Descriptor: | (3R)-1-(2-{[1-(pyrimidin-5-yl)cyclopropyl]amino}pyrimidine-5-carbonyl)piperidine-3-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
8QG4
 
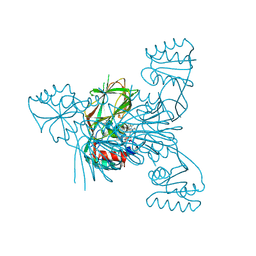 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
8QGO
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative | | Descriptor: | 1-[[(2~{R},3~{S},4~{R},5~{R})-5-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-ethyl-urea, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G, Lionne, C. | | Deposit date: | 2023-09-05 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a di-adenosine derivative
To Be Published
|
|
5MX7
 
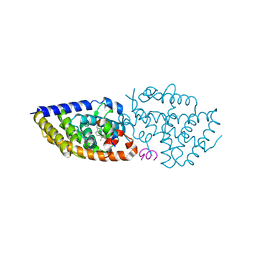 | | 1a,20S-dihydroxyvitamin D3 VDR complex | | Descriptor: | 1a,20S-dihydroxyvitamin D3, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2017-01-21 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 1 alpha,20S-Dihydroxyvitamin D3 Interacts with Vitamin D Receptor: Crystal Structure and Route of Chemical Synthesis.
Sci Rep, 7, 2017
|
|
6R5H
 
 | | Major aspartyl peptidase 1 from C. neoformans | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Krystufek, R, Sacha, P, Brynda, J, Konvalinka, J. | | Deposit date: | 2019-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery.
J.Med.Chem., 64, 2021
|
|
3CSO
 
 | | HCV Polymerase in complex with a 1,5 Benzodiazepine inhibitor | | Descriptor: | (11S)-10-acetyl-11-[4-(benzyloxy)-3-chlorophenyl]-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, RNA-directed RNA polymerase | | Authors: | Nyanguile, O. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | 1,5-benzodiazepines, a novel class of hepatitis C virus polymerase nonnucleoside inhibitors.
Antimicrob.Agents Chemother., 52, 2008
|
|
8E5N
 
 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 10 | | Descriptor: | 1-{[(3S,4S)-3-(3-fluorophenyl)-4-{[4-(1,3,4-triethyl-1H-pyrazol-5-yl)piperidin-1-yl]methyl}pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
9NLP
 
 | | HIV-1 Reverse Transcriptase with New Non-Nucleoside Reverse Transcriptase Inhibitor 12126065 | | Descriptor: | 4-({5-amino-1-[6-(2-cyanoethyl)naphthalene-1-sulfonyl]-1H-1,2,4-triazol-3-yl}amino)-2-chlorobenzonitrile, Reverse transcriptase p51 subunit, Reverse transcriptase p66 subunit | | Authors: | Young, M.A, Lane, T.R, Raman, R, Nelson, J.A.E, Riabova, O, Kazakova, E, Monakhova, N, Tsedilin, A, Rees, S.D, Quinnell, D, Chang, G, Ekins, S. | | Deposit date: | 2025-03-03 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Cryo-EM Structure of HIV-1 Reverse Transcriptase with N -Phenyl-1-(phenylsulfonyl)-1 H -1,2,4-triazol-3-amine: A New HIV-1 Non-nucleoside Inhibitor.
Acs Infect Dis., 11, 2025
|
|
8E5M
 
 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 6 | | Descriptor: | 1-{[(3S,4S)-3-({4-[2-(4-fluorobenzene-1-sulfonyl)ethyl]piperidin-1-yl}methyl)-4-(3-fluorophenyl)pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
3FUC
 
 | | Recombinant calf purine nucleoside phosphorylase in a binary complex with multisubstrate analogue inhibitor 9-(5',5'-difluoro-5'-phosphonopentyl)-9-deazaguanine structure in a new space group with one full trimer in the asymmetric unit | | Descriptor: | AZIDE ION, MAGNESIUM ION, Purine nucleoside phosphorylase, ... | | Authors: | Bochtler, M, Breer, K, Bzowska, A, Chojnowski, G, Hashimoto, M, Hikishima, S, Narczyk, M, Wielgus-Kutrowska, B, Yokomatsu, T. | | Deposit date: | 2009-01-14 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 A resolution crystal structure of recombinant PNP in complex with a pM multisubstrate analogue inhibitor bearing one feature of the postulated transition state.
Biochem.Biophys.Res.Commun., 391, 2010
|
|
8DX8
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 2-chloro-6-fluorophenethylamine at the 415 site | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-chloro-6-fluorophenyl)ethan-1-amine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
5TGD
 
 | |
7CUY
 
 | | Crystal structure of Primo-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Low molecular weight phosphotyrosine protein phosphatase 1 | | Authors: | Lee, H.S, Kim, S.J, Ku, B. | | Deposit date: | 2020-08-25 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Structural and Biochemical Characterization of the Two Drosophila Low Molecular Weight-Protein Tyrosine Phosphatases DARP and Primo-1.
Mol.Cells, 43, 2020
|
|
8DXI
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol at multiple sites | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
8DX3
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 3-bromobenzylamine in the thumb subdomain | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-bromophenyl)methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
3EKT
 
 | | Crystal Structure of the inhibitor Darunavir (DRV) in complex with a multi-drug resistant HIV-1 protease variant (L10F/G48V/I54V/V64I/V82A) (Refer: FLAP+ in citation.) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
7N05
 
 | |
3FX5
 
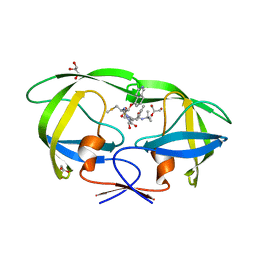 | | Structure of HIV-1 Protease in Complex with Potent Inhibitor KNI-272 Determined by High Resolution X-ray Crystallography | | Descriptor: | (4R)-N-tert-butyl-3-[(2S,3S)-2-hydroxy-3-({N-[(isoquinolin-5-yloxy)acetyl]-S-methyl-L-cysteinyl}amino)-4-phenylbutanoyl]-1,3-thiazolidine-4-carboxamide, GLYCEROL, protease | | Authors: | Adachi, M, Ohhara, T, Tamada, T, Okazaki, N, Kuroki, R. | | Deposit date: | 2009-01-20 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Structure of HIV-1 protease in complex with potent inhibitor KNI-272 determined by high-resolution X-ray and neutron crystallography.
Proc.Natl.Acad.Sci.USA, 2009
|
|
3G7Q
 
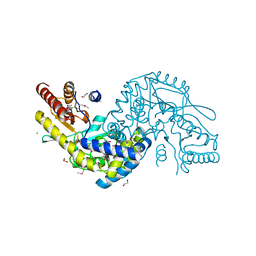 | |
5AHC
 
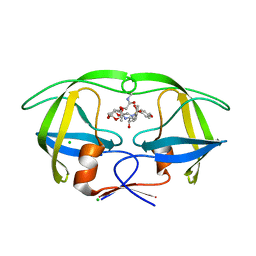 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aR)-4-[2-(methylamino)-2-oxoethoxy]hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
5AH7
 
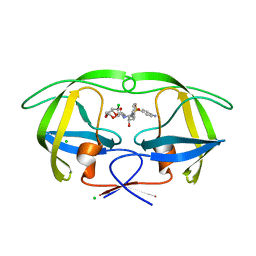 | | Disubstituted bis-THF moieties as new P2 ligands in non-peptidal HIV- 1 Protease Inhibitors (II) | | Descriptor: | (3R,3aS,4R,6aS)-4-chlorohexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, PROTEASE | | Authors: | Hohlfeld, K, Wegner, J.K, Kesteleyn, B, Linclau, B, Unge, J. | | Deposit date: | 2015-02-05 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Disubstituted Bis-Thf Moieties as New P2 Ligands in Non-Peptidal HIV-1 Protease Inhibitors (II).
J.Med.Chem., 58, 2015
|
|
4BUH
 
 | | Human IgE against the major allergen Bet v 1 - Crystal structure of clone M0418 scFv | | Descriptor: | 1,2-ETHANEDIOL, CLONE M0418 SCFV, DI(HYDROXYETHYL)ETHER | | Authors: | Davies, A.M, Levin, M, Lilljekvist, M, Carlsson, F, Gould, H.J, Sutton, B.J, Ohlin, M. | | Deposit date: | 2013-06-20 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Human Ige Against the Major Allergen Bet V 1 - Defining an Epitope with Limited Cross-Reactivity between Different Pr-10 Family Proteins
Clin.Exp.Allergy, 44, 2014
|
|
3DB6
 
 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 902 | | Descriptor: | 1-[5-methyl-2-(trifluoromethyl)furan-3-yl]-3-[(2Z)-5-(2-{[6-(1H-1,2,4-triazol-3-ylamino)pyrimidin-4-yl]amino}ethyl)-1,3-thiazol-2(3H)-ylidene]urea, Polo-like kinase 1 | | Authors: | Elling, R.A, Fucini, R.V, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-30 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Brachydanio rerio Polo-like kinase 1 (Plk1) catalytic domain in complex with an extended inhibitor targeting the adaptive pocket of the enzyme.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
4XYC
 
 | | NANOMOLAR INHIBITORS OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE 1: SYNTHESIS, BIOLOGICAL EVALUATION AND X-RAY CRYSTALLOGRAPHIC STUDIES | | Descriptor: | 9-phenyl-4H-imidazo[1,2-a]indeno[1,2-e]pyrazin-4-one, Glutamine synthetase 1 | | Authors: | Couturier, C, Silve, S, Morales, R, Ppessegue, B, Llopart, S, Nair, A, Bauer, A, Scheiper, B, poeverlein, c, Ganzhorn, A, Lagrange, S, Bacque, E. | | Deposit date: | 2015-02-02 | | Release date: | 2015-03-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
