6UDR
 
 | | S2 symmetric peptide design number 3 crystal form 1, Lurch | | Descriptor: | S2-3, Lurch crystal form 1 | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
7UKL
 
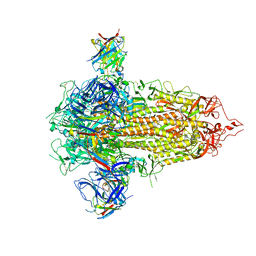 | |
7UUG
 
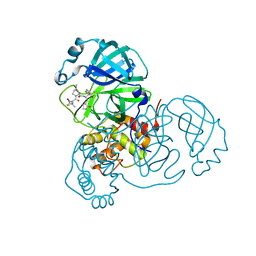 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUP
 
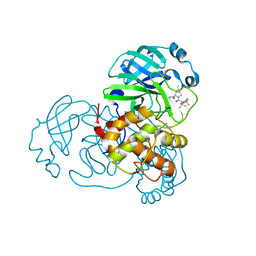 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
5APZ
 
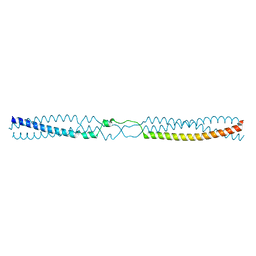 | | Thermosinus carboxydivorans Nor1 Tcar0761 residues 68-101 and 191-211 fused to GCN4 adaptors | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, NOR1 TCAR0761 | | Authors: | Hartmann, M.D, Deiss, S, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
6UD9
 
 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5AMY
 
 | |
6UCX
 
 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-02 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
5APP
 
 | | Actinobacillus actinomycetemcomitans OMP100 residues 133-198 fused to GCN4 adaptors | | Descriptor: | CHLORIDE ION, General control protein GCN4,GENERAL CONTROL PROTEIN GCN4, OUTER MEMBRANE PROTEIN 100,General control protein GCN4 | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
4UCW
 
 | | Structure of the T18V small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Threonine Stabilizes the Nic and Nir Catalytic Intermediates of [Nife]-Hydrogenase.
J.Biol.Chem., 290, 2015
|
|
5FHQ
 
 | |
5APX
 
 | | Sequence MATKDDIAN inserted between GCN4 adaptors - Structure T9(6) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PHOSPHATE ION | | Authors: | Hartmann, M.D, Mendler, C.T, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2015-09-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | alpha / beta coiled coils.
Elife, 5, 2016
|
|
5APT
 
 | |
5APS
 
 | |
5FZ3
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 3,6-Dihydroxybenzonorbornane (N08776b) (ligand modelled based on PANDDA event map) | | Descriptor: | (1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalene-5,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Pearce, N, Talon, R, Collins, P, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N08776B
To be Published
|
|
5E78
 
 | | Crystal structure of P450 BM3 heme domain variant complexed with Co(III)Sep | | Descriptor: | 1,3,6,8,10,13,16,19-octaazabicyclo[6.6.6]icosane, Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, ... | | Authors: | Panneerselvm, S, Shehzad, A, Bocola, M, Mueller-Dieckmann, J, Schwaneberg, U. | | Deposit date: | 2015-10-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic insights into a cobalt (III) sepulchrate based alternative cofactor system of P450 BM3 monooxygenase.
Biochim. Biophys. Acta, 1866, 2018
|
|
5DXJ
 
 | | Crystal structure of CARM1 and sinefungin | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, SINEFUNGIN | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2015-09-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Insights into Ternary Complex Formation of Human CARM1 with Various Substrates.
Acs Chem.Biol., 11, 2016
|
|
5HDI
 
 | | Structural characterization of CYP144A1, a Mycobacterium tuberculosis cytochrome P450 | | Descriptor: | Cytochrome P450 144, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Chenge, J, Driscoll, M.D, McLean, K.J, Munro, A.W, Leys, D. | | Deposit date: | 2016-01-05 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of CYP144A1 - a cytochrome P450 enzyme expressed from alternative transcripts in Mycobacterium tuberculosis.
Sci Rep, 6, 2016
|
|
2HW1
 
 | | Crystal structure of human ketohexokinase complexed to different sugar molecules | | Descriptor: | GLYCEROL, Ketohexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Trinh, C.H, Asipu, A, Bonthron, D.T, Phillips, S.E.V. | | Deposit date: | 2006-07-31 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of alternatively spliced isoforms of human ketohexokinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
7Z78
 
 | | Crystal structure of compound 4 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | 4-bromanyl-7-cyclopentyl-9-piperidin-4-yl-benzimidazolo[1,2-a]quinazolin-5-one, Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo.
Nat Commun, 13, 2022
|
|
2HQQ
 
 | | Crystal structure of human ketohexokinase complexed to different sugar molecules | | Descriptor: | Ketohexokinase, SULFATE ION | | Authors: | Trinh, C.H, Asipu, A, Bonthron, D.T, Phillips, S.E.V. | | Deposit date: | 2006-07-19 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of alternatively spliced isoforms of human ketohexokinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1DEF
 
 | |
2JJP
 
 | | Structure of cytochrome P450 EryK in complex with inhibitor ketoconazole (KC) | | Descriptor: | 1-ACETYL-4-(4-{[(2S,4R)-2-(2,4-DICHLOROPHENYL)-2-(1H-IMIDAZOL-1-YLMETHYL)-1,3-DIOXOLAN-4-YL]METHOXY}PHENYL)PIPERAZINE, CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Azole Drugs Trap Cytochrome P450 Eryk in Alternative Conformational States.
Biochemistry, 49, 2010
|
|
4WX7
 
 | | Crystal structure of adenovirus 8 protease with a nitrile inhibitor | | Descriptor: | 3-[2-(3,5-dichlorophenyl)-2-methylpropanoyl]-N-(2-{[(2Z)-2-iminoethyl]amino}-2-oxoethyl)-4-methoxybenzamide, PVI, Protease | | Authors: | Grosche, P, Sirockin, F, Mac Sweeney, A, Ramage, P, Erbel, P, Melkko, S, Bernardi, A, Hughes, N, Ellis, D, Combrink, K, Jarousse, N, Altmann, E. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2015-01-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design and optimization of potent inhibitors of the adenoviral protease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
