4C8C
 
 | | mouse ZNRF3 ectodomain crystal form III | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8T
 
 | | Xenopus ZNRF3 ectodomain crystal form I | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8A
 
 | | mouse ZNRF3 ectodomain crystal form II | | Descriptor: | E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8V
 
 | | Xenopus RSPO2 Fu1-Fu2 crystal form I | | Descriptor: | R-SPONDIN-2 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C9U
 
 | |
4CKB
 
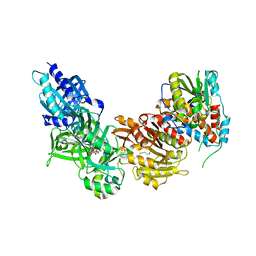 | | Vaccinia virus capping enzyme complexed with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, ... | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4C9E
 
 | |
4PKV
 
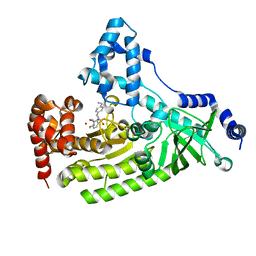 | | Anthrax toxin lethal factor with bound small molecule inhibitor 16 | | Descriptor: | Lethal factor, N~2~-[4-(aminomethyl)benzyl]-N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-D-alaninamide, ZINC ION | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PKQ
 
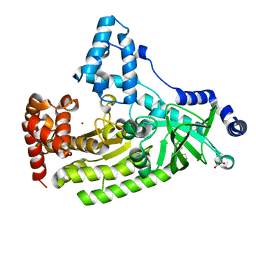 | | Anthrax toxin lethal factor with bound zinc | | Descriptor: | 1,2-ETHANEDIOL, Lethal factor, TRIETHYLENE GLYCOL, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C8U
 
 | | Xenopus ZNRF3 ectodomain crystal form II | | Descriptor: | BROMIDE ION, E3 UBIQUITIN-PROTEIN LIGASE ZNRF3 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4C8W
 
 | | Xenopus RSPO2 Fu1-Fu2 crystal form II | | Descriptor: | R-SPONDIN-2 | | Authors: | Zebisch, M, Jones, E.Y. | | Deposit date: | 2013-10-01 | | Release date: | 2013-11-20 | | Last modified: | 2013-11-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Molecular Basis of Znrf3/Rnf43 Transmembrane Ubiquitin Ligase Inhibition by the Wnt Agonist R-Spondin.
Nat.Commun., 4, 2013
|
|
4PKT
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 13 | | Descriptor: | Lethal factor, N~2~-[(4-fluoro-3-methylphenyl)sulfonyl]-N-hydroxy-N~2~-(4-nitrobenzyl)-D-alaninamide, ZINC ION | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2N21
 
 | | Solution structure of complex between DNA G-quadruplex and G-quadruplex recognition domain of RHAU | | Descriptor: | ATP-dependent RNA helicase DHX36, DNA (5'-D(*TP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*TP*GP*GP*GP*T)-3') | | Authors: | Heddi, B, Cheong, V.V, Martadinata, H, Phan, A.T. | | Deposit date: | 2015-04-25 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4C8P
 
 | |
4C9V
 
 | |
4CKE
 
 | | Vaccinia virus capping enzyme complexed with SAH in P1 form | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-03 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
4CKC
 
 | | Vaccinia virus capping enzyme complexed with SAH (monoclinic form) | | Descriptor: | MRNA-CAPPING ENZYME CATALYTIC SUBUNIT, MRNA-CAPPING ENZYME REGULATORY SUBUNIT, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kyrieleis, O.J.P, Chang, J, de la Pena, M, Shuman, S, Cusack, S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Vaccinia Virus Mrna Capping Enzyme Provides Insights Into the Mechanism and Evolution of the Capping Apparatus.
Structure, 22, 2014
|
|
3PFQ
 
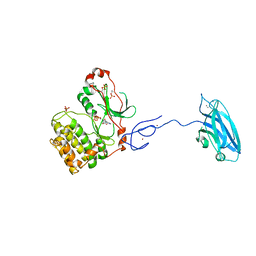 | | Crystal Structure and Allosteric Activation of Protein Kinase C beta II | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase C beta type, ... | | Authors: | Leonard, T.A, Rozycki, B, Saidi, L.F, Hummer, G, Hurley, J.H. | | Deposit date: | 2010-10-28 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Cell(Cambridge,Mass.), 144, 2011
|
|
2MAJ
 
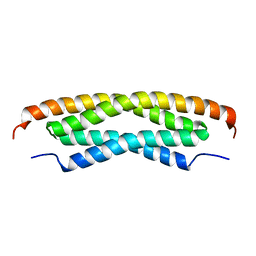 | |
2MAK
 
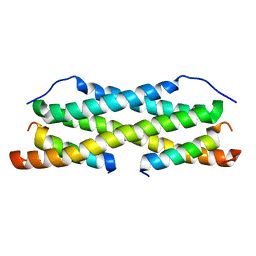 | |
2PV9
 
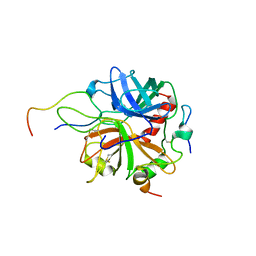 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 4, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PUX
 
 | | Crystal structure of murine thrombin in complex with the extracellular fragment of murine PAR3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 3, Thrombin heavy chain, ... | | Authors: | Bah, A, Chen, Z, Bush-Pelc, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2007-05-09 | | Release date: | 2007-07-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of murine thrombin in complex with the extracellular fragments of murine protease-activated receptors PAR3 and PAR4.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3LHT
 
 | | Crystal structure of the mutant V201F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LI1
 
 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-23 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3LLD
 
 | | Crystal structure of the mutant S127G of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-01-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
