7PVQ
 
 | |
8PZ9
 
 | | Crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 Analog 55 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{R},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,8,9,9~{a}-hexahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2023-08-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
5E73
 
 | | Crystal Structure of BAZ2B bromodomain in complex with acetylindole compound UZH47 | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2B, N-(1-acetyl-1H-indol-3-yl)-N-(5-hydroxy-2-methylphenyl)acetamide | | Authors: | Lolli, G, Spiliotopoulos, D, Unzue, A, Nevado, C, Caflisch, A. | | Deposit date: | 2015-10-11 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The "Gatekeeper" Residue Influences the Mode of Binding of Acetyl Indoles to Bromodomains.
J. Med. Chem., 59, 2016
|
|
6NCG
 
 | | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor | | Descriptor: | 4-[6-amino-5-(3,5-difluoro-4-hydroxyphenyl)pyridin-3-yl]benzene-1-sulfonamide, SULFATE ION, Serine/threonine-protein kinase VRK2 | | Authors: | dos Reis, C.V, Chiodi, C.G, de Souza, G.P, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Counago, R.M, Massirer, K.B, Elkins, J.M, Arruda, P, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-11 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Human Vaccinia-related kinase 2 (VRK-2) bound to pyridin-benzenesulfonamide inhibitor
To Be Published
|
|
7N3S
 
 | |
5DUW
 
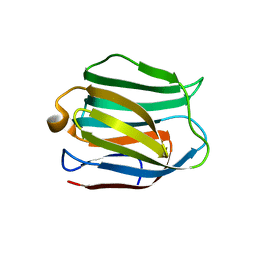 | |
6NCQ
 
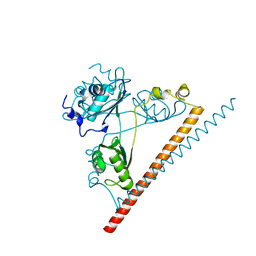 | | The dimerization domain of human SFPQ in space group C2221 | | Descriptor: | Splicing factor, proline- and glutamine-rich | | Authors: | Lee, M. | | Deposit date: | 2018-12-12 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A new crystal structure and small-angle X-ray scattering analysis of the homodimer of human SFPQ.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8T8I
 
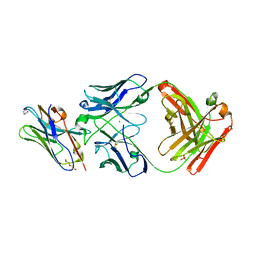 | | Structure of VHH-Fab complex with engineered Elbow FNQIKG, Crystal Kappa and SER substitutions | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fab heavy chain, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-22 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
7W44
 
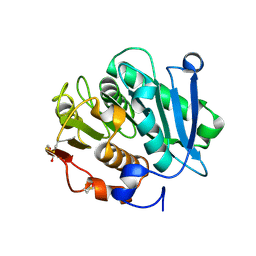 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7YOR
 
 | | Recj2 and dCMP complex from Methanocaldococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, [5-(4-azanyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-furan-2-yl]methyl dihydrogen phosphate, archaeal nuclease RecJ2 | | Authors: | Wang, W.W, Liu, X.P. | | Deposit date: | 2022-08-02 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The structure of nuclease RecJ2 from Methanocaldococcus jannaschii
To Be Published
|
|
8SMJ
 
 | |
7PVX
 
 | |
7PW2
 
 | |
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
6NCW
 
 | |
7WRK
 
 | |
5E7O
 
 | | Crystal structure of the perchlorate reductase PcrAB mutant W461E of PcrA from Azospira suillum PS | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO reductase family type II enzyme, ... | | Authors: | Tsai, C.-L, Tainer, J.A. | | Deposit date: | 2015-10-12 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Perchlorate Reductase Is Distinguished by Active Site Aromatic Gate Residues.
J.Biol.Chem., 291, 2016
|
|
6NDJ
 
 | | Crystal structure of human NLRP6 PYD domain with MBP fusion | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, NACHT, LRR and PYD domains-containing protein 6 chimera | | Authors: | Shen, C, Fu, T.M, Wu, H. | | Deposit date: | 2018-12-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Molecular mechanism for NLRP6 inflammasome assembly and activation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7N3R
 
 | |
6NID
 
 | |
7W2E
 
 | |
5VC8
 
 | | Crystal structure of the WHSC1 PWWP1 domain | | Descriptor: | DNA (5'-D(P*CP*TP*(DN))-3'), Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ... | | Authors: | Qin, S, Tempel, W, Dong, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Biochem.Biophys.Res.Commun., 569, 2021
|
|
8STS
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (Y181C, V106A) varient in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)-4-fluorophenoxy)-7-fluoro-2-naphthonitrile (JLJ636), a non-nucleoside inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-7-fluoronaphthalene-2-carbonitrile, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Hollander, K, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-05-11 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Exploring novel HIV-1 reverse transcriptase inhibitors with drug-resistant mutants: A double mutant surprise.
Protein Sci., 32, 2023
|
|
7YMU
 
 | | Structure of Alcohol dehydrogenase from [Candida] glabrata | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sun, Z.W, Liu, Y.F, Xu, G.C, Ni, Y. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rationla design of CgADH from candida glarata for asymmetric reduction of azacycolne.
To Be Published
|
|
7PJM
 
 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and Ca2+/2-Oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
