8QPC
 
 | |
8CP0
 
 | | Structure of the catalytic domain of P. vivax Sub1 (trigonal crystal form) | | Descriptor: | CALCIUM ION, subtilisin | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.251 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7XIH
 
 | |
8COZ
 
 | | Structure of the catalytic domain of P. vivax Sub1 (triclinic crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, SULFATE ION, ... | | Authors: | Martinez, M, Bouillon, A, Batista, F, Alzari, P.M, Barale, J.C, Haouz, A. | | Deposit date: | 2023-03-01 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.438 Å) | | Cite: | 3D structures of the Plasmodium vivax subtilisin-like drug target SUB1 reveal conformational changes to accommodate a substrate-derived alpha-ketoamide inhibitor.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8QXX
 
 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
7TZA
 
 | | Structure of PQS Response Protein PqsE in complex with N-(4-(3-neopentylureido)phenyl)-1H-indazole-7-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, N-{4-[(2,2-dimethylpropyl)carbamamido]phenyl}-1H-indazole-7-carboxamide, ... | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PqsE Active Site as a Target for Small Molecule Antimicrobial Agents against Pseudomonas aeruginosa.
Biochemistry, 61, 2022
|
|
6N6N
 
 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
7XIG
 
 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) and spermine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
8QLT
 
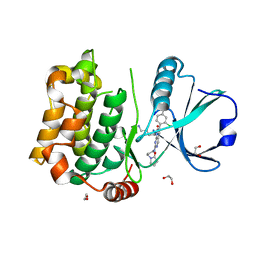 | | Human MST3 (STK24) kinase in complex with inhibitor MR30 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-[3-(2-oxidanylidenepyrrolidin-1-yl)propylamino]pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6VJ3
 
 | | Carbonic Anhydrase II in complex with pyrimidine-based inhibitor | | Descriptor: | 2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-N-(4-sulfamoylphenyl)acetamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Lomelino, C.L, McKenna, R. | | Deposit date: | 2020-01-14 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Inclusion of a 5-fluorouracil moiety in nitrogenous bases derivatives as human carbonic anhydrase IX and XII inhibitors produced a targeted action against MDA-MB-231 and T47D breast cancer cells.
Eur.J.Med.Chem., 190, 2020
|
|
6BIV
 
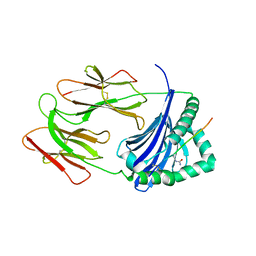 | | HLA-DRB1 in complex with citrullinated LL37 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
7X0F
 
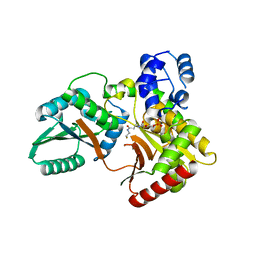 | |
8QXW
 
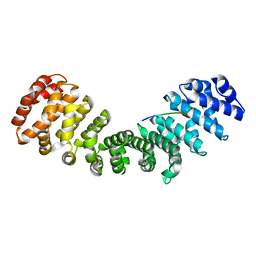 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
7P47
 
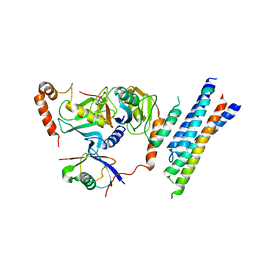 | | Structure of the E3 ligase Smc5/Nse2 in complex with Ubc9-SUMO thioester mimetic | | Descriptor: | E3 SUMO-protein ligase MMS21, SUMO-conjugating enzyme UBC9, Structural maintenance of chromosomes protein 5, ... | | Authors: | Lascorz, J, Varejao, N, Reverter, D. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Nat Commun, 12, 2021
|
|
6POT
 
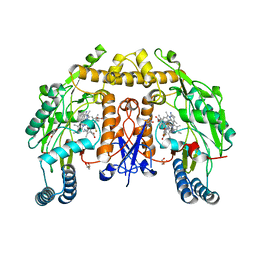 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(3-(Aminomethyl)-4-(thiazol-5-ylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-{3-(aminomethyl)-4-[(1,3-thiazol-5-yl)methoxy]phenyl}-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8QLS
 
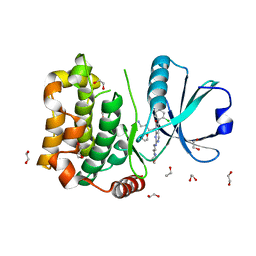 | | Human MST3 (STK24) kinase in complex with inhibitor MR26 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2-chloranyl-4-(6-methylpyridin-2-yl)phenyl]-2-(3-morpholin-4-ylpropylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Development of Selective Pyrido[2,3- d ]pyrimidin-7(8 H )-one-Based Mammalian STE20-Like (MST3/4) Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
6W4Z
 
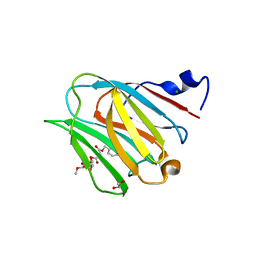 | | Galectin-8N terminal domain in complex with Methyl 3-O-[3-O-benzyloxy]-malonyl-beta-D-galactopyranoside | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Galectin-8, ... | | Authors: | Patel, B, Kishor, C, Blanchard, H. | | Deposit date: | 2020-03-12 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rational Design and Synthesis of Methyl-beta-d-galactomalonyl Phenyl Esters as Potent Galectin-8 N Antagonists.
J.Med.Chem., 63, 2020
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
7XIF
 
 | | Crystal structure of the aminopropyltransferase, SpeE from hyperthermophilic crenarchaeon, Pyrobaculum calidifontis in complex with 5'-methylthioadenosine (MTA) alone or together with spermidine or thermospermine | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, N-(3-AMINO-PROPYL)-N-(5-AMINOPROPYL)-1,4-DIAMINOBUTANE, Polyamine aminopropyltransferase, ... | | Authors: | Mizohata, E, Yasuda, Y. | | Deposit date: | 2022-04-13 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Substrate Specificity of an Aminopropyltransferase and the Biosynthesis Pathway of Polyamines in the Hyperthermophilic Crenarchaeon Pyrobaculum calidifontis.
Catalysts, 12, 2022
|
|
7U6G
 
 | |
6PP0
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-(cyclobutylmethoxy)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-(cyclobutylmethoxy)phenyl]-4-methylquinolin-2-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-05 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8QLQ
 
 | | Human MST3 (STK24) kinase in complex with macrocyclic inhibitor JA310 | | Descriptor: | 1,2-ETHANEDIOL, Serine/threonine-protein kinase 24, macrocyclic inhibitor | | Authors: | Balourdas, D.I, Amrhein, J.A, Hanke, T, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Synthesis of Pyrazole-Based Macrocycles Leads to a Highly Selective Inhibitor for MST3.
J.Med.Chem., 67, 2024
|
|
6PIC
 
 | |
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
6W5E
 
 | | Class D beta-lactamase BSU-2 | | Descriptor: | 1,2-ETHANEDIOL, BSU-2 beta-lactamase, MALONATE ION | | Authors: | Smith, C.A, Vakulenko, S.B, Stewart, N.K, Toth, M. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | A surface loop modulates activity of the Bacillus class D beta-lactamases.
J.Struct.Biol., 211, 2020
|
|
