7XVN
 
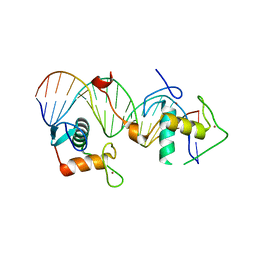 | | Structural basis for DNA recognition feature of retinoid-related orphan receptors | | Descriptor: | DNA (5'-D(P*CP*AP*TP*GP*AP*CP*CP*TP*AP*CP*TP*GP*AP*CP*CP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*GP*GP*TP*CP*AP*GP*TP*AP*GP*GP*TP*CP*AP*TP*G)-3'), Nuclear receptor ROR-gamma, ... | | Authors: | Chen, Y, Jiang, L. | | Deposit date: | 2022-05-24 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural characterization of the DNA binding mechanism of retinoic acid-related orphan receptor gamma.
Structure, 32, 2024
|
|
4RVE
 
 | | THE CRYSTAL STRUCTURE OF ECORV ENDONUCLEASE AND OF ITS COMPLEXES WITH COGNATE AND NON-COGNATE DNA SEGMENTS | | Descriptor: | DNA (5'-D(*GP*GP*GP*AP*TP*AP*TP*CP*CP*C)-3'), PROTEIN (ECO RV (E.C.3.1.21.4)) | | Authors: | Winkler, F.K, Banner, D.W, Oefner, C, Tsernoglou, D, Brown, R.S, Heathman, S.P, Bryan, R.K, Martin, P.D, Petratos, K, Wilso, K.S. | | Deposit date: | 1993-02-18 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments.
EMBO J., 12, 1993
|
|
3VH0
 
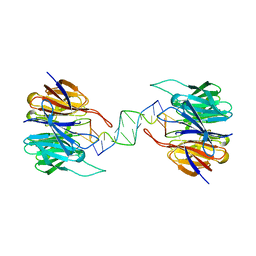 | | Crystal structure of E. coli YncE complexed with DNA | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*TP*AP*CP*TP*CP*AP*G)-3'), Uncharacterized protein YncE | | Authors: | Kagawa, W, Sagawa, T, Niki, H, Kurumizaka, H. | | Deposit date: | 2011-08-23 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the DNA-binding activity of the bacterial beta-propeller protein YncE
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5HAW
 
 | |
7UYO
 
 | |
7UYN
 
 | |
7UYP
 
 | |
2VAN
 
 | | Nucleotidyl Transfer Mechanism of Mismatched dNTP Incorporation by DNA Polymerase b by Structural and Kinetic Analyses | | Descriptor: | DNA POLYMERSE BETA | | Authors: | Chan, H, Chou, C, Tang, K, Niebuhr, M, Tung, C, Tsai, M. | | Deposit date: | 2007-09-03 | | Release date: | 2008-04-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mismatched Dntp Incorporation by DNA Polymerase Beta Does not Proceed Via Globally Different Conformational Pathways.
Nucleic Acids Res., 36, 2008
|
|
7JSF
 
 | | Adeno-Associated Virus Helicase domain Heptamer with ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7JSI
 
 | | Adeno-Associated Virus 2 Rep68 HD Hexamer-ssDNA with ATPgS | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*T)-3'), Protein Rep68 | | Authors: | Escalante, C.R. | | Deposit date: | 2020-08-14 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.01 Å) | | Cite: | The Cryo-EM structure of AAV2 Rep68 in complex with ssDNA reveals a malleable AAA+ machine that can switch between oligomeric states.
Nucleic Acids Res., 48, 2020
|
|
7KZS
 
 | |
7O56
 
 | | X-ray Structure of Interferon Regulatory Factor 4 DNA binding domain bound to an interferon-stimulated response element solved by Phosphorus and Sulphur SAD methods | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*AP*GP*AP*AP*AP*CP*CP*GP*AP*AP*AP*GP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*AP*CP*TP*TP*TP*CP*GP*GP*TP*TP*TP*CP*TP*TP*TP*TP*AP*T)-3'), Interferon regulatory factor 4 | | Authors: | El Omari, K, Agnarelli, A, Duman, R, Wagner, A, Mancini, E.J. | | Deposit date: | 2021-04-07 | | Release date: | 2021-05-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phosphorus and sulfur SAD phasing of the nucleic acid-bound DNA-binding domain of interferon regulatory factor 4.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4OWX
 
 | | Structural basis of SOSS1 in complex with a 12nt ssDNA | | Descriptor: | DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), Integrator complex subunit 3, SOSS complex subunit B1 | | Authors: | Ren, W, Sun, Q, Tang, X, Song, H. | | Deposit date: | 2014-02-04 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA.
Cell Rep, 6, 2014
|
|
5F55
 
 | | Structure of RecJ complexed with DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*GP*TP*AP*CP*GP*CP*TP*AP*GP*GP*C)-3'), MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hua, Y, Zhao, Y, Cheng, K. | | Deposit date: | 2015-12-04 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for DNA 5 -end resection by RecJ
Elife, 5, 2016
|
|
5IUD
 
 | |
1D12
 
 | | STRUCTURAL COMPARISON OF ANTICANCER DRUG-DNA COMPLEXES. ADRIAMYCIN AND DAUNOMYCIN | | Descriptor: | DNA (5'-D(*CP*GP*AP*TP*CP*G)-3'), DOXORUBICIN, SODIUM ION, ... | | Authors: | Frederick, C.A, Williams, L.D, Ughetto, G, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J. | | Deposit date: | 1989-10-20 | | Release date: | 1990-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural comparison of anticancer drug-DNA complexes: adriamycin and daunomycin.
Biochemistry, 29, 1990
|
|
3U2B
 
 | | Structure of the Sox4 HMG domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*AP*CP*AP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*TP*GP*TP*CP*CP*TP*GP*G)-3'), Transcription factor SOX-4 | | Authors: | Jauch, R, Ng, C.K.L, Kolatkar, P.R. | | Deposit date: | 2011-10-03 | | Release date: | 2011-12-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | The crystal structure of the Sox4 HMG domain-DNA complex suggests a mechanism for positional interdependence in DNA recognition
Biochem.J., 443, 2012
|
|
8QXW
 
 | | HCMV DNA polymerase processivity factor UL44 unphosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
8QXX
 
 | | HCMV DNA polymerase processivity factor UL44 phosphorylated NLS 410-433 bound to mouse importin alpha 2 | | Descriptor: | DNA polymerase processivity factor, Importin subunit alpha-1 | | Authors: | Cross, E.M, Marin, O, Ariawan, D, Aragao, D, Cozza, G, Di Iorio, E, Forwood, J.K, Alvisi, G. | | Deposit date: | 2023-10-25 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Febs Lett., 598, 2024
|
|
1D29
 
 | |
7SLB
 
 | | [T:Ag+:T--pH 8.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7SLE
 
 | | [T:Ag+:T--pH 10] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.39 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7SLD
 
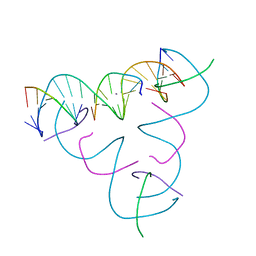 | | [T:Ag+:T--pH 9.5] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.13 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7SLC
 
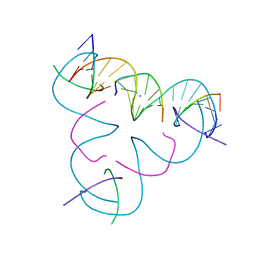 | | [T:Ag+:T--pH 9] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*TP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*TP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
7SLM
 
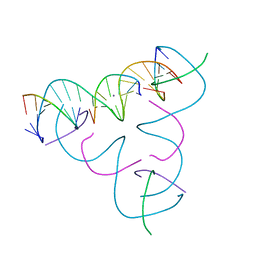 | | [U:Ag+:U--pH 10] Metal-mediated DNA base pair in tensegrity triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*UP*TP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*AP*UP*AP*CP*A)-3'), DNA (5'-D(P*CP*TP*GP*AP*TP*GP*T)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P. | | Deposit date: | 2021-10-24 | | Release date: | 2022-10-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Heterobimetallic Base Pair Programming in Designer 3D DNA Crystals.
J.Am.Chem.Soc., 145, 2023
|
|
