6DIS
 
 | | Crystal structure of HCV NS3/4A protease in complex with P4-1 (AJ-71) | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5, 16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6- yl]carbamate, GLYCEROL, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6PA1
 
 | | Killer cell immunoglobulin-like receptor 2DL2 in complex with HLA-C*07:02 | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Moradi, S, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C.
Nat Commun, 12, 2021
|
|
6PAG
 
 | | Killer cell immunoglobulin-like receptor 2DL3 in complex with HLA-C*07:02 | | Descriptor: | ARG-TYR-ARG-PRO-GLY-THR-VAL-ALA-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Moradi, S, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2019-06-11 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C.
Nat Commun, 12, 2021
|
|
8DZL
 
 | | Structure of the K39Q mutant of rat somatic Cytochrome c at 1.36A | | Descriptor: | Cytochrome c, somatic, HEME C | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
6DIQ
 
 | |
6DIW
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-1 (AJ-71) | | Descriptor: | 1,1,1-trifluoro-2-methylpropan-2-yl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5, 16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6- yl]carbamate, GLYCEROL, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6XNK
 
 | |
9HUA
 
 | | Glycosyltransferase C from the Limosilactobacillus reuteri accessory secretion system. Complex with UDP. | | Descriptor: | Glucosyltransferase 3, URIDINE-5'-DIPHOSPHATE | | Authors: | Asworth, G.J, Griffiths, R, Juge, N, Dong, C.J, Hemmings, A.M. | | Deposit date: | 2024-12-21 | | Release date: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the strain-dependent UDP-sugar specificity of glycosyltransferase C from the Limosilactobacillus reuteri accessory secretion system
To Be Published
|
|
5KZP
 
 | |
6Y57
 
 | | Structure of human ribosome in hybrid-PRE state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-25 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
6VD9
 
 | |
6GC5
 
 | |
8WCT
 
 | |
8WCN
 
 | | Cryo-EM structure of PAO1-ImcA with GMPCPP | | Descriptor: | Diguanylate cyclase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Zhan, X.L, Zhang, K, Wang, C.C, Fan, Q, Tang, X.J, Zhang, X, Wang, K, Fu, Y, Liang, H.H. | | Deposit date: | 2023-09-13 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A c-di-GMP signaling module controls responses to iron in Pseudomonas aeruginosa.
Nat Commun, 15, 2024
|
|
5LXV
 
 | |
7UN8
 
 | | SfSTING with c-di-GMP single fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
7UN9
 
 | | SfSTING with c-di-GMP double fiber | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Morehouse, B.R, Yip, M.C.J, Keszei, A.F.A, McNamara-Bordewick, N.K, Shao, S, Kranzusch, P.J. | | Deposit date: | 2022-04-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of an active bacterial TIR-STING filament complex.
Nature, 608, 2022
|
|
5D9G
 
 | | Crystal structure of TIPRL, TOR signaling pathway regulator-like, in complex with peptide | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PHOSPHATE ION, ... | | Authors: | Scorsato, V, Sandy, J, Brandao-Neto, J, Pereira, H.M, Smetana, J.H.C, Aparicio, R. | | Deposit date: | 2015-08-18 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human Tip41 orthologue, TIPRL, reveals a novel fold and a binding site for the PP2Ac C-terminus.
Sci Rep, 6, 2016
|
|
9CZ2
 
 | |
6SX4
 
 | |
9DF3
 
 | | EGFR Exon20 insertion mutant SVD bound with (S)-3-((3-chloro-2-methoxyphenyl)amino)-2-(3-((tetrahydrofuran-2-yl)methoxy)pyridin-4-yl)-1,5,6,7-tetrahydro-4H-pyrrolo[3,2-c]pyridin-4-one | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-[3-[[(2S)-oxolan-2-yl]methoxy]pyridin-4-yl]-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, Epidermal growth factor receptor, ... | | Authors: | Hilbert, B.J, Brooijmans, N, Pagliarini, R.A, Milgram, B.C. | | Deposit date: | 2024-08-29 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | STX-721, a Covalent EGFR/HER2 Exon 20 Inhibitor, Utilizes Exon 20-Mutant Dynamic Protein States and Achieves Unique Mutant Selectivity Across Human Cancer Models.
Clin.Cancer Res., 2025
|
|
9EHY
 
 | | X-ray crystal structure of ADC-33 beta-lactamase in complex with ceftazidime in acyl and product forms | | Descriptor: | (2R)-2-[(R)-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}(carboxy)methyl]-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, ACYLATED CEFTAZIDIME, Beta-lactamase | | Authors: | Powers, R.A, Wallar, B.J, Jarvis, H.R, Ziegler, Z.X, June, C.M. | | Deposit date: | 2024-11-25 | | Release date: | 2025-05-14 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Resistance to oxyimino-cephalosporins conferred by an alternative mechanism of hydrolysis by the Acinetobacter -derived cephalosporinase-33 (ADC-33), a class C beta-lactamase present in carbapenem-resistant Acinetobacter baumannii (CR Ab ).
Mbio, 16, 2025
|
|
8QRI
 
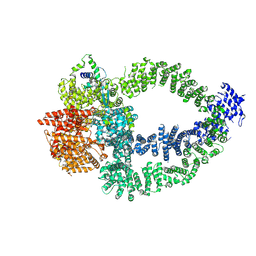 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 635, 2024
|
|
6XFS
 
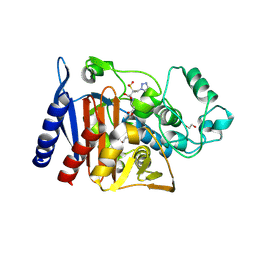 | | Class C beta-lactamase from Escherichia coli in complex with Tazobactam | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Class C beta-lactamase from Escherichia coli in complex with Tazobactam
To Be Published
|
|
6N5W
 
 | |
