5F1X
 
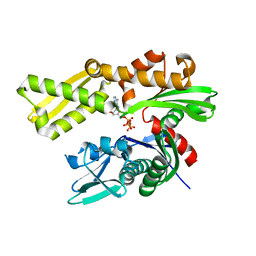 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EY4
 
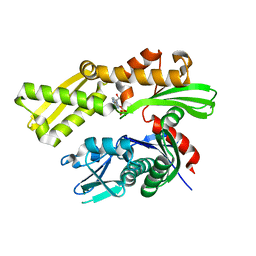 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F0X
 
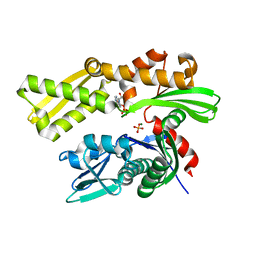 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ADP and inorganic phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 78 kDa glucose-regulated protein, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EX5
 
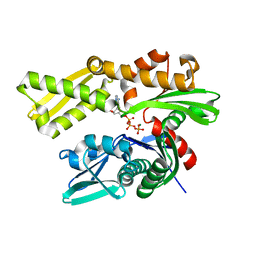 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ADP and inorganic phosphate | | Descriptor: | 7-deazaadenosine-5'-diphosphate, 78 kDa glucose-regulated protein, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5FPD
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand pyrazine-2-carboxamide (AT513) in an alternate binding site. | | Descriptor: | HEAT SHOCK-RELATED 70KDA PROTEIN 2, PYRAZINE-2-CARBOXAMIDE | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3FE1
 
 | | Crystal structure of the human 70kDa heat shock protein 6 (Hsp70B') ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 6, ... | | Authors: | Wisniewska, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
5GJJ
 
 | |
5FPN
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 3,5-dimethyl-1H-pyrazole-4-carboxylic acid (AT9084) in an alternate binding site. | | Descriptor: | 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, HEAT SHOCK-RELATED 70 KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPE
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 1H-1,2,4-triazol-3-amine (AT485) in an alternate binding site. | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3D2F
 
 | | Crystal structure of a complex of Sse1p and Hsp70 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1, ... | | Authors: | Polier, S, Bracher, A. | | Deposit date: | 2008-05-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding.
Cell(Cambridge,Mass.), 133, 2008
|
|
5FPM
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3CQX
 
 | | Chaperone Complex | | Descriptor: | BAG family molecular chaperone regulator 2, Heat shock cognate 71 kDa protein, SODIUM ION, ... | | Authors: | Xu, Z, Nix, J.C, Misra, S. | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2
Nat.Struct.Mol.Biol., 15, 2008
|
|
5EVZ
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
3DPO
 
 | |
3DPP
 
 | |
3DPQ
 
 | |
3GDQ
 
 | | Crystal structure of the human 70kDa heat shock protein 1-like ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Heat shock 70 kDa protein 1-like, ... | | Authors: | Wisniewska, M, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Lehtio, L, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-02-24 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
4HY9
 
 | |
1NGD
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
4HYB
 
 | |
4IO8
 
 | | Crystal structure of human HSP70 complexed with 4-{(2R,3S,4R)-5-[(R)-6-Amino-8-(3,4-dichloro-benzylamino)-purin-9-yl]-3,4-dihydroxy-tetrahydro-furan-2-ylmethoxymethyl}-benzonitrile | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Heat shock 70kDa protein 1A variant | | Authors: | Musil, D, Scholz, S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Functional analysis of hsp70 inhibitors.
Plos One, 8, 2013
|
|
1NGJ
 
 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
4JWE
 
 | |
4JWC
 
 | |
7KRW
 
 | |
