5X2Q
 
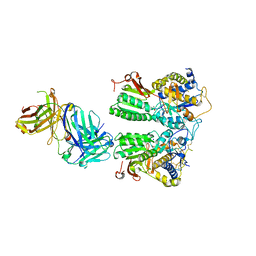 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
2FY2
 
 | |
5I9I
 
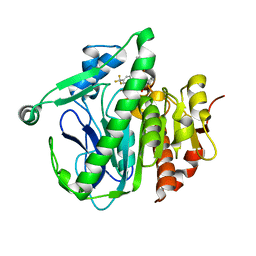 | | Crystal structure of LP_PLA2 in complex with Darapladib | | Descriptor: | N-[2-(diethylamino)ethyl]-2-{2-[(4-fluorobenzyl)sulfanyl]-4-oxo-4,5,6,7-tetrahydro-1H-cyclopenta[d]pyrimidin-1-yl}-N-{[ 4'-(trifluoromethyl)biphenyl-4-yl]methyl}acetamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Characterization of Protein-Ligand Interactions Formed between Lipoprotein-Associated Phospholipase A2 and Inhibitors
J.Med.Chem., 59, 2016
|
|
2FY5
 
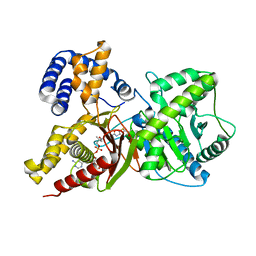 | | Structures of ligand bound human choline acetyltransferase provide insight into regulation of acetylcholine synthesis | | Descriptor: | Choline O-acetyltransferase, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-4-HYDROXY-3-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL (3R)-3-HYDROXY-2,2-DIMETHYL-4-OXO-4-{[3-OXO-3-({2-[(2-OXOPROPYL)THIO]ETHYL}AMINO)PROPYL]AMINO}BUTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Kim, A.R, Rylett, R.J, Shilton, B.H. | | Deposit date: | 2006-02-07 | | Release date: | 2006-12-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate binding and catalytic mechanism of human choline acetyltransferase.
Biochemistry, 45, 2006
|
|
8G66
 
 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
5X2O
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
1IRA
 
 | | COMPLEX OF THE INTERLEUKIN-1 RECEPTOR WITH THE INTERLEUKIN-1 RECEPTOR ANTAGONIST (IL1RA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-1 RECEPTOR, INTERLEUKIN-1 RECEPTOR ANTAGONIST | | Authors: | Schreuder, H.A, Tardif, C, Tramp-Kalmeyer, S, Soffientini, A, Sarubbi, E, Akeson, A, Bowlin, T, Yanofsky, S, Barrett, R.W. | | Deposit date: | 1998-04-09 | | Release date: | 1998-06-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist.
Nature, 386, 1997
|
|
4XR8
 
 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
2QO1
 
 | | 2.6 Angstrom Crystal Structure of the Complex Between 11-(decyldithiocarbonyloxy)-undecanoic acid and Mycobacterium Tuberculosis FabH. | | Descriptor: | 11-[(MERCAPTOCARBONYL)OXY]UNDECANOIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase 3, DECANE-1-THIOL | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-07-19 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Separate Entrance and Exit Portals for Ligand Traffic in Mycobacterium tuberculosis FabH
Chem.Biol., 15, 2008
|
|
7PG0
 
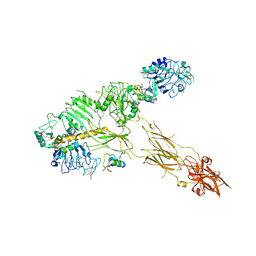 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin with visible ddm micelle, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG2
 
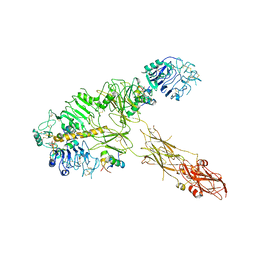 | | Low resolution Cryo-EM structure of full-length insulin receptor bound to 3 insulin, conf 1 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG4
 
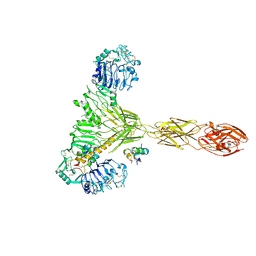 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 2 insulin, conf 3 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (9.1 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
7PG3
 
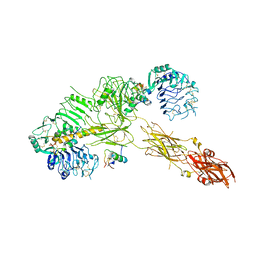 | | Low resolution Cryo-EM structure of the full-length insulin receptor bound to 3 insulin, conf 2 | | Descriptor: | Insulin, Isoform Short of Insulin receptor | | Authors: | Nielsen, J.A, Slaaby, R, Boesen, T, Hummelshoj, T, Brandt, J, Schluckebier, G, Nissen, P. | | Deposit date: | 2021-08-12 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Investigations of Full-Length Insulin Receptor Dynamics and Signalling.
J.Mol.Biol., 434, 2022
|
|
5JG9
 
 | |
4GB0
 
 | |
1CAH
 
 | |
6N4X
 
 | | Metabotropic Glutamate Receptor 5 Apo Form Ligand Binding Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Metabotropic glutamate receptor 5 | | Authors: | Koehl, A, Hu, H, Feng, D, Sun, B, Weis, W.I, Skiniotis, G.S, Mathiesen, J.M, Kobilka, B.K. | | Deposit date: | 2018-11-20 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insights into the activation of metabotropic glutamate receptors.
Nature, 566, 2019
|
|
3BUX
 
 | | Crystal structure of c-Cbl-TKB domain complexed with its binding motif in c-Met | | Descriptor: | 13-meric peptide from Hepatocyte growth factor receptor, E3 ubiquitin-protein ligase CBL | | Authors: | Ng, C, Jackson, R.A, Buschdorf, J.P, Sun, Q, Guy, G.R, Sivaraman, J. | | Deposit date: | 2008-01-03 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates
Embo J., 27, 2008
|
|
7PI0
 
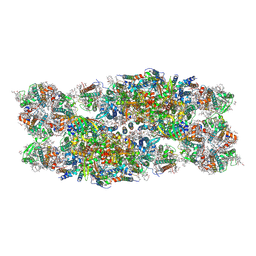 | | Unstacked compact Dunaliella PSII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Caspy, I, Fadeeva, M, Mazor, Y, Nelson, N. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-17 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Structure of Dunaliella Photosystem II reveals conformational flexibility of stacked and unstacked supercomplexes.
Elife, 12, 2023
|
|
7EA9
 
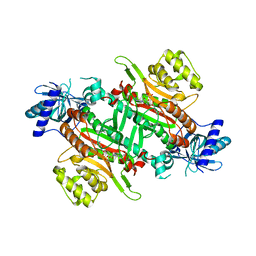 | | Crystal Structure of human lysyl-tRNA synthetase Y145H mutant | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, GLYCEROL, Lysine--tRNA ligase | | Authors: | Wu, S, Hei, Z, Zheng, L, Zhou, J, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2021-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analyses of a human lysyl-tRNA synthetase mutant associated with autosomal recessive nonsyndromic hearing impairment.
Biochem.Biophys.Res.Commun., 554, 2021
|
|
6BKW
 
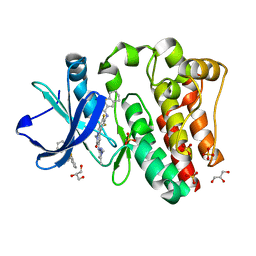 | | BTK complex with compound 12 | | Descriptor: | GLYCEROL, N-(3-{5-[(1,5-dimethyl-1H-pyrazol-3-yl)amino]-1-methyl-6-oxo-1,6-dihydropyridazin-3-yl}-2,6-difluorophenyl)-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, SULFATE ION, ... | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-07 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
6BIK
 
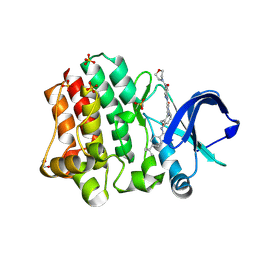 | | BTK complex with compound 7 | | Descriptor: | 4-tert-butyl-N-[2-(hydroxymethyl)-3-(1-methyl-5-{[5-(morpholine-4-carbonyl)pyridin-2-yl]amino}-6-oxo-1,6-dihydropyridazin-3-yl)phenyl]benzamide, GLYCEROL, SULFATE ION, ... | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-02 | | Release date: | 2018-11-07 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
4M4N
 
 | |
6BKH
 
 | | BTK complex with compound 11 | | Descriptor: | N-[2-(hydroxymethyl)-3-{5-[(5-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl)amino]-6-oxo-1,6-dihydropyridazin-3-yl}phenyl]-1-benzothiophene-2-carboxamide, SULFATE ION, Tyrosine-protein kinase BTK | | Authors: | Kiefer, J.R, Eigenbrot, C, Yu, C.L. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-07 | | Last modified: | 2019-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Water molecules in protein-ligand interfaces. Evaluation of software tools and SAR comparison.
J. Comput. Aided Mol. Des., 33, 2019
|
|
1HJ4
 
 | |
