8UCY
 
 | |
8PWH
 
 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
8UGY
 
 | | Serotonin 1E receptor (5-HT1eR)-Gi1 Complex bound with Mianserin | | Descriptor: | 5-hydroxytryptamine receptor 1E, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zilberg, G, Warren, A.L, Wacker, D. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural insights into the unexpected agonism of tetracyclic antidepressants at serotonin receptors 5-HT 1e R and 5-HT 1F R.
Sci Adv, 10, 2024
|
|
8UD0
 
 | |
8UCZ
 
 | |
8V9M
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (R)-3-amino-5,5-difluorocyclohex-1-ene-1-carboxylic acid. | | Descriptor: | 3-fluoro-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]benzoic acid, GLYCEROL, Ornithine aminotransferase, ... | | Authors: | Vargas, A.L, Devitt, A, Kaley, N, Silverman, R, Liu, D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, and Mechanistic Studies of ( R )-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase.
Acs Chem.Biol., 19, 2024
|
|
6Z1R
 
 | | bovine ATP synthase F1-peripheral stalk domain, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z1U
 
 | | bovine ATP synthase F1c8-peripheral stalk domain, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6RZX
 
 | | Carbonic Anhydrase CAIX mimic in complex with inhibitor FBSA | | Descriptor: | 1,1,2,2,3,3,4,4,4-nonakis(fluoranyl)butane-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Kugler, M, Brynda, J, Rezacova, P. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Identification of Novel Carbonic Anhydrase IX Inhibitors Using High-Throughput Screening of Pooled Compound Libraries by DNA-Linked Inhibitor Antibody Assay (DIANA).
SLAS Discov, 25, 2020
|
|
8VDD
 
 | | Crystal structure of Proinsulin C-peptide bound to HLA-DQ8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, MHC class II HLA-DQ-alpha chain, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 2024
|
|
8VD0
 
 | | Human TCR ET650-4 in complex with DQ8-InsC8-15-IAPP2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Hybrid insulin peptide (HIP; InsC8-15-IAPP74-80),MHC class II HLA-DQ-beta-1 chimera, ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-14 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 2024
|
|
8VDU
 
 | | Crystal structure of hybrid insulin peptide (InsC8-15-IAPP74-80) bound to HLA-DQ8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Hybrid insulin peptide (HIP; InsC8-15-IAPP74-80), ... | | Authors: | Tran, T.M, Lim, J.J, Loh, T.Y, Mannering, I.S, Rossjohn, J, Reid, H.H. | | Deposit date: | 2023-12-17 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A structural basis of T cell cross-reactivity to native and spliced self-antigens presented by HLA-DQ8.
J.Biol.Chem., 2024
|
|
7LVS
 
 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
8W6A
 
 | |
8W6B
 
 | |
3BTS
 
 | |
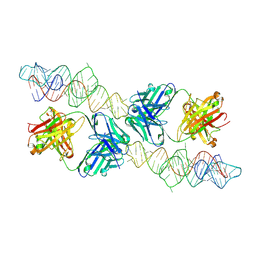
8VM9
 
 | |
8VMA
 
 | |
8VM8
 
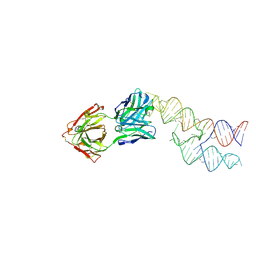 | |
8VMB
 
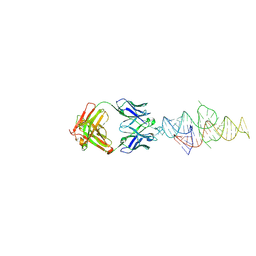 | |
1HBB
 
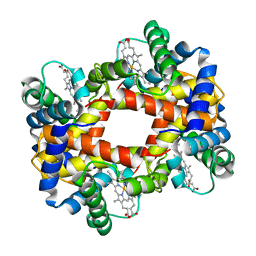 | |
1HBA
 
 | |
5BUI
 
 | | ERK2 complexed with 2-pyridiyl tetrahydroazaindazole | | Descriptor: | 3-(4-fluorophenyl)-5-(pyridin-2-yl)-4,5,6,7-tetrahydro-2H-pyrazolo[4,3-c]pyridine, Mitogen-activated protein kinase 1, NICKEL (II) ION | | Authors: | Bellamacina, C.R, Shu, W, Bussiere, D.E, Bagdanoff, J.T. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Ligand efficient tetrahydro-pyrazolopyridines as inhibitors of ERK2 kinase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1 Y108F MUTANT | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE, S-HEXYLGLUTATHIONE | | Authors: | Oakley, A, Rossjohn, J, Parker, M. | | Deposit date: | 1997-01-20 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme.
Biochemistry, 36, 1997
|
|
4LBT
 
 | | Endothiapepsin in complex with 100mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, DIMETHYL SULFOXIDE, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Winquist, J, Klebe, G. | | Deposit date: | 2013-06-21 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
