7KOX
 
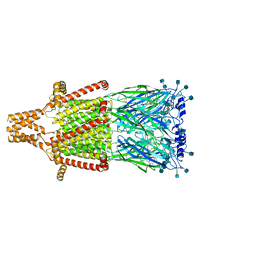 | | Alpha-7 nicotinic acetylcholine receptor bound to epibatidine and PNU-120596 in the activated state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Baxter, L, Cabuco, R, Borek, D, Sine, S. | | Deposit date: | 2020-11-10 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KOO
 
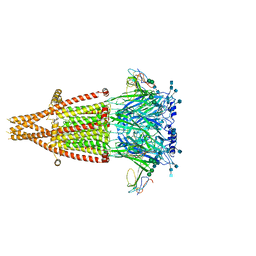 | | Alpha-7 nicotinic acetylcholine receptor bound to alpha-bungarotoxin in a resting state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-bungarotoxin isoform V31, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
7KOQ
 
 | | Alpha-7 nicotinic acetylcholine receptor bound to epibatidine in a desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Noviello, C.M, Hibbs, R.E, Gharpure, A, Mukhtasimova, N, Cabuco, R, Baxter, L, Borek, D, Sine, S. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and gating mechanism of the alpha 7 nicotinic acetylcholine receptor.
Cell, 184, 2021
|
|
6MRC
 
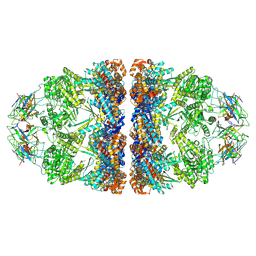 | | ADP-bound human mitochondrial Hsp60-Hsp10 football complex | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Gomez-Llorente, Y, Jebara, F, Patra, M, Malik, R, Nissemblat, S, Azem, A, Hirsch, J.A, Ubarretxena-Belandia, I. | | Deposit date: | 2018-10-12 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for active single and double ring complexes in human mitochondrial Hsp60-Hsp10 chaperonin.
Nat Commun, 11, 2020
|
|
2IJX
 
 | |
1K4Z
 
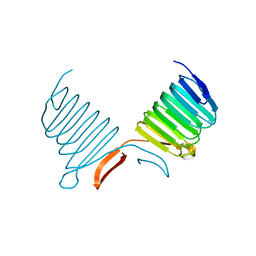 | | C-terminal Domain of Cyclase Associated Protein | | Descriptor: | Adenylyl Cyclase-Associated Protein | | Authors: | Rozwarski, D.A, Fedorov, A.A, Dodatko, T, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-09 | | Release date: | 2002-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein
Biochemistry, 43, 2004
|
|
6MU2
 
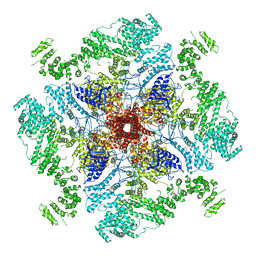 | | Structure of full-length IP3R1 channel in the Apo-state | | Descriptor: | Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Serysheva, I.I, Fan, G, Baker, M.R, Wang, Z, Seryshev, A, Ludtke, S.J, Baker, M.L. | | Deposit date: | 2018-10-22 | | Release date: | 2018-12-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM reveals ligand induced allostery underlying InsP3R channel gating.
Cell Res., 28, 2018
|
|
6B8P
 
 | |
6VAC
 
 | | Mouse retromer (VPS26/VPS35/VPS29) heterotrimer | | Descriptor: | Vacuolar protein sorting-associated protein 26A, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | Authors: | Kendall, A.K, Jackson, L.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes.
Structure, 28, 2020
|
|
7KL5
 
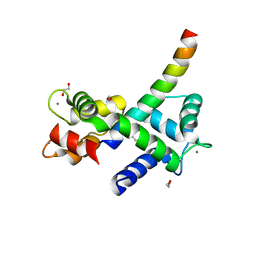 | | Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues: Phe4246 to Val4271 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calmodulin-1, ... | | Authors: | Fisher, A.J, Ames, J.B, Yu, Q. | | Deposit date: | 2020-10-29 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Biochemistry, 60, 2021
|
|
2IPU
 
 | |
7QEP
 
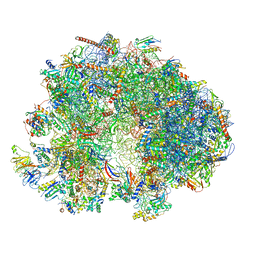 | | Cryo-EM structure of the ribosome from Encephalitozoon cuniculi | | Descriptor: | 18S ribosomal RNA, 40S RIBOSOMAL PROTEIN S10, 40S RIBOSOMAL PROTEIN S11, ... | | Authors: | Nicholson, D, Ranson, N.A, Melnikov, S.V. | | Deposit date: | 2021-12-03 | | Release date: | 2022-02-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Adaptation to genome decay in the structure of the smallest eukaryotic ribosome
Nat Commun, 13, 2022
|
|
6BC8
 
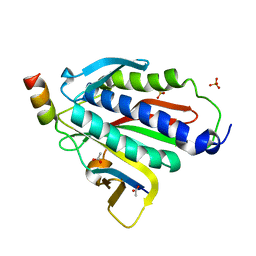 | | Crystal structure of Rev7-R124A/Rev3-RBM2 (residues 1988-2014) complex | | Descriptor: | ACETATE ION, DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B, ... | | Authors: | Rizzo, A.A, Hao, B, Li, Y, Korzhnev, D.M. | | Deposit date: | 2017-10-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Rev7 dimerization is important for assembly and function of the Rev1/Pol zeta translesion synthesis complex.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5JD0
 
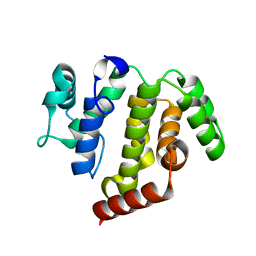 | | crystal structure of ARAP3 RhoGAP domain | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3 | | Authors: | Bao, H, Li, F, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
6AX5
 
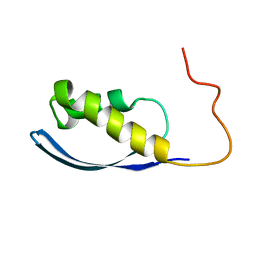 | | RPT1 region of INI1/SNF5/SMARCB1_HUMAN - SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Girvin, M.E, Cahill, S.M, Harris, R, Cowburn, D, Spira, M, Wu, X, Prakash, R, Bernowitz, M, Almo, S.C, Kalpana, G.V. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | INI1/SMARCB1 Rpt1 domain mimics TAR RNA in binding to integrase to facilitate HIV-1 replication.
Nat Commun, 12, 2021
|
|
6Y78
 
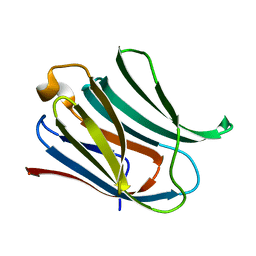 | | Structure of galectin-3C in complex with lactose determined by serial crystallography using a silicon nitride membrane support | | Descriptor: | Galectin-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Hakansson, M, Welin, M, Shilova, A, Kovacic, R, Mueller, U, Logan, D.T. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Current status and future opportunities for serial crystallography at MAX IV Laboratory.
J.Synchrotron Radiat., 27, 2020
|
|
1K8F
 
 | | CRYSTAL STRUCTURE OF THE HUMAN C-TERMINAL CAP1-ADENYLYL CYCLASE ASSOCIATED PROTEIN | | Descriptor: | ADENYLYL CYCLASE-ASSOCIATED PROTEIN | | Authors: | Patskovsky, Y.V, Chance, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-10-24 | | Release date: | 2003-07-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the actin binding domain of the cyclase-associated protein.
Biochemistry, 43, 2004
|
|
6N2M
 
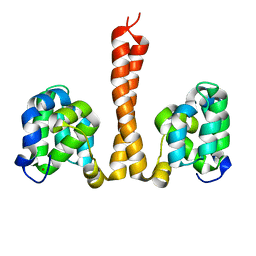 | |
6BHN
 
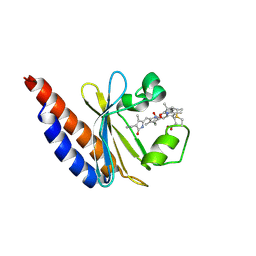 | | Red Light-Absorbing State of NpR6012g4, a Red/Green Cyanobacteriochrome | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with phytochrome sensor, PHYCOCYANOBILIN | | Authors: | Yu, Q, Lim, S, Rockwell, N.C, Martin, S.S, Lagarias, J.C, Ames, J.B. | | Deposit date: | 2017-10-31 | | Release date: | 2018-04-18 | | Last modified: | 2019-12-04 | | Method: | SOLUTION NMR | | Cite: | Correlating structural and photochemical heterogeneity in cyanobacteriochrome NpR6012g4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5MRW
 
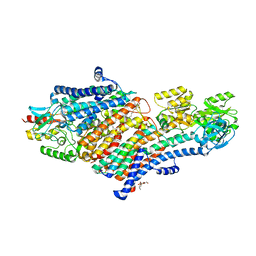 | | Structure of the KdpFABC complex | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Huang, C, Pedersen, B.P, Stokes, D.L. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the potassium-importing KdpFABC membrane complex.
Nature, 546, 2017
|
|
5G1C
 
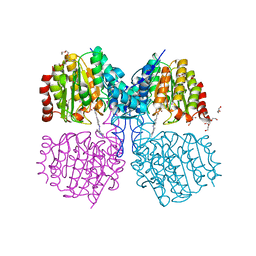 | | Structure of HDAC like protein from Bordetella Alcaligenes bound the photoswitchable pyrazole Inhibitor CEW395 | | Descriptor: | (2E)-N-hydroxy-3-{4-[(E)-(1,3,5-trimethyl-1H-pyrazol-4-yl)diazenyl]phenyl}prop-2-enamide, DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Toward Photopharmacological Antimicrobial Chemotherapy Using Photoswitchable Amidohydrolase Inhibitors.
ACS Infect Dis, 3, 2017
|
|
1I9E
 
 | | TCR DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTOTOXIC TCELL VALPHA DOMAIN | | Authors: | Rudolph, M.G, Huang, M, Teyton, L, Wilson, I.A. | | Deposit date: | 2001-03-19 | | Release date: | 2001-12-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an isolated V(alpha) domain of the 2C T-cell receptor.
J.Mol.Biol., 314, 2001
|
|
6VFF
 
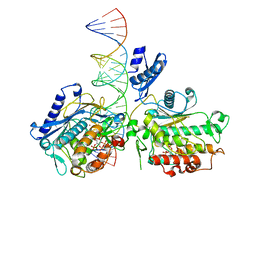 | | Dimer of Human Adenosine Deaminase Acting on dsRNA (ADAR2) mutant E488Q bound to dsRNA sequence derived from human GLI1 gene | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5-R(*GP*CP*UP*CP*GP*CP*GP*AP*UP*GP*CP*UP*(8AZ)P*GP*AP*GP*GP*GP*CP* UP*CP*UP*GP*AP*UP*AP*GP*CP*UP*AP*CP*G)-3), ... | | Authors: | Thuy-boun, A.S, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-01-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asymmetric dimerization of adenosine deaminase acting on RNA facilitates substrate recognition.
Nucleic Acids Res., 48, 2020
|
|
1I38
 
 | |
7WZS
 
 | |
