8DK4
 
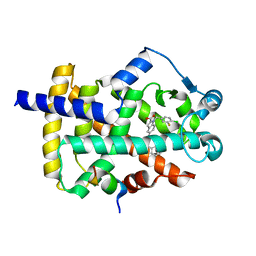 | | Peroxisome proliferator-activated receptor gamma in complex with VSP-51-2 | | Descriptor: | 5-(benzylcarbamoyl)-1-[(4-chloro-3-fluorophenyl)methyl]-1H-indole-2-carboxylic acid, Peroxisome proliferator-activated receptor gamma, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha | | Authors: | Ma, L, Zhou, X.E, Suino-Powell, K, Hou, N, Zhou, Z, Luo, J, Xu, H.E, Yi, W. | | Deposit date: | 2022-07-02 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of VSP-51-2 as the Novel and Safe PPAR gamma Modulator: Structure-Based Design, Biological Validation and Crystal Analysis
To Be Published
|
|
7TQ6
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 13d | | Descriptor: | (1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[(1R,2R)-2-propylcyclopropyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7W24
 
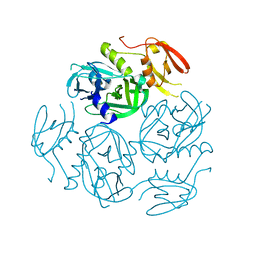 | | Structure of the M. tuberculosis HtrA N383A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
6P90
 
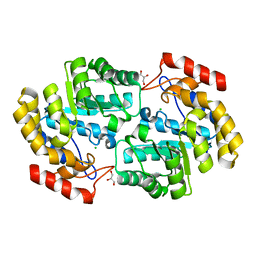 | | Crystal structure of PaDHDPS2-H56Q mutant | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate synthase, CHLORIDE ION, GLYCEROL | | Authors: | Impey, R.E, Panjikar, S, Hall, C.J, Bock, L.J, Sutton, J.M, Perugini, M.A, Soares da Costa, T.P. | | Deposit date: | 2019-06-08 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation.
Febs J., 287, 2020
|
|
8DHK
 
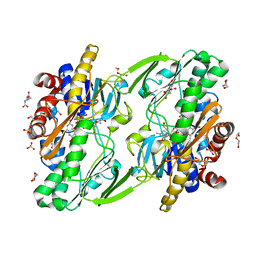 | |
4ZYP
 
 | | Crystal Structure of Motavizumab and Quaternary-Specific RSV-Neutralizing Human Antibody AM14 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | AM14 antibody Fab heavy chain, AM14 antibody light chain, Fusion glycoprotein F0,Fibritin, ... | | Authors: | Gilman, M.S.A, McLellan, J.S. | | Deposit date: | 2015-05-21 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Characterization of a Prefusion-Specific Antibody That Recognizes a Quaternary, Cleavage-Dependent Epitope on the RSV Fusion Glycoprotein.
Plos Pathog., 11, 2015
|
|
6UV6
 
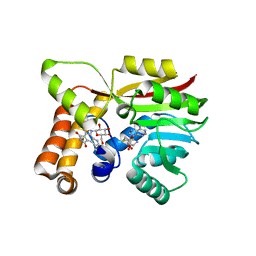 | | AtmM with bound rebeccamycin analogue | | Descriptor: | 12-beta-D-glucopyranosyl-12,13-dihydro-5H-indolo[2,3-a]pyrrolo[3,4-c]carbazole-5,7(6H)-dione, D-glucose O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Alvarado, S.K, Wang, Z, Miller, M.D, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of AtmM Bound with Glycosylated Indolocarbazole
To Be Published
|
|
6MJC
 
 | |
8DAE
 
 | |
6P99
 
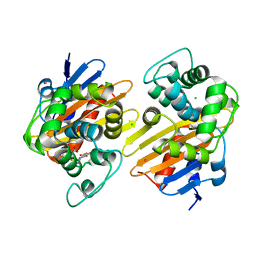 | | OXA-48 carbapanemase, ertapenem complex | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CADMIUM ION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Insights into the Mechanism of Carbapenemase Activity of the OXA-48 beta-Lactamase.
Antimicrob.Agents Chemother., 63, 2019
|
|
7W25
 
 | | Structure of the M. tuberculosis HtrA S413A mutant | | Descriptor: | Probable serine protease HtrA1 | | Authors: | Gupta, A.K, Gopal, B. | | Deposit date: | 2021-11-21 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Allosteric Determinants in High Temperature Requirement A Enzymes Are Conserved and Regulate the Population of Active Conformations.
Acs Chem.Biol., 18, 2023
|
|
7TM8
 
 | |
4ZZM
 
 | | Human ERK2 in complex with an irreversible inhibitor | | Descriptor: | 7-ethylsulfonyl-N-(oxan-4-yl)-6,8-dihydro-5H-pyrido[3,4-d]pyrimidin-2-amine, MITOGEN-ACTIVATED PROTEIN KINASE 1, SULFATE ION | | Authors: | Ward, R.A, Colclough, N, Challinor, M, Debreczeni, J.E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, James, M, Jones, C.D, Jones, C.R, Renshaw, J, Roberts, K, Snow, L, Tonge, M, Yeung, K. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-27 | | Last modified: | 2015-08-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-Guided Design of Highly Selective and Potent Covalent Inhibitors of Erk1/2.
J.Med.Chem., 58, 2015
|
|
7P1L
 
 | | The MARK3 Kinase Domain Bound To AA-CS-1-008 | | Descriptor: | 1,2-ETHANEDIOL, 5-Bromo-4-N-[2-(1H-imidazol-5-yl)ethyl]-2-N-[3-(morpholin-4-ylmethyl)phenyl]pyrimidine-2,4-diamine, MAP/microtubule affinity-regulating kinase 3 | | Authors: | Dederer, V, Preuss, F, Chatterjee, D, Vlassova, A, Mathea, S, Axtman, A, Knapp, S. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Identification of Pyrimidine-Based Lead Compounds for Understudied Kinases Implicated in Driving Neurodegeneration.
J.Med.Chem., 65, 2022
|
|
8DGR
 
 | |
7TMB
 
 | |
6AUT
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(Dimethylamino)propyl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6PQC
 
 | | Structure of cefotaxime-CDD-1 beta-lactamase complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of CDD-1, the intrinsic class D beta-lactamase from the pathogenic Gram-positive bacterium Clostridioides difficile, and its complex with cefotaxime.
J.Struct.Biol., 208, 2019
|
|
6MKH
 
 | | Crystal structure of pencillin binding protein 4 (PBP4) from Enterococcus faecalis in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, PHOSPHATE ION, pencillin binding protein 4 (PBP4) | | Authors: | D'Andrea, E.D, Moon, T.M, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
7TM7
 
 | |
6B2E
 
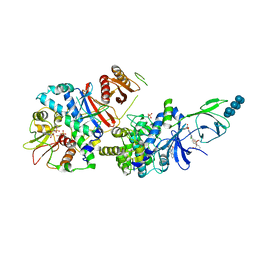 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | Deposit date: | 2017-09-19 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
8Q29
 
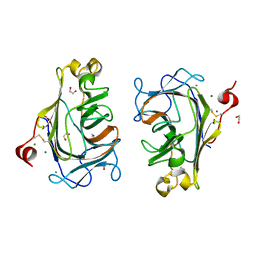 | |
8Q1W
 
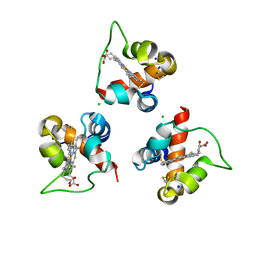 | |
8DE5
 
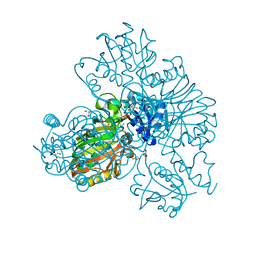 | | Structure of glyceraldehyde-3-phosphate dehydrogenase from Paracoccidioides lutzii | | Descriptor: | D-galactonic acid, GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, ... | | Authors: | Hernandez-Prieto, J.H, Martini, V.P, Iulek, J. | | Deposit date: | 2022-06-19 | | Release date: | 2023-06-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of glyceraldehyde-3-phosphate dehydrogenase from Paracoccidioides lutzii in complex with an aldonic sugar acid.
Biochimie, 218, 2023
|
|
7TM5
 
 | |
