1C5U
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PROTEIN (TRYPSIN), ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5R
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C9A
 
 | | SOLUTION STRUCTURE OF NEUROMEDIN B | | Descriptor: | NEUROMEDIN B | | Authors: | Lee, S, Kim, Y. | | Deposit date: | 1999-08-01 | | Release date: | 1999-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of neuromedin B by (1)H nuclear magnetic resonance spectroscopy.
FEBS Lett., 460, 1999
|
|
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
7FX8
 
 | | Crystal Structure of human FABP4 in complex with 4-(2-methylpropyl)-2-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, i.e. SMILES c1(c2c(C3=NN=NN3)c(CC(C)C)c3c(n2)CCCCC3)ccccc1 with IC50=0.0809837 microM | | Descriptor: | (3M)-4-(2-methylpropyl)-2-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
8FE2
 
 | | Structure of J-PKAc chimera complexed with Aplithianine A | | Descriptor: | 6-[(6M)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-9H-purine, DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Du, L, Wilson, B.A.P, Li, N, Dalilian, M, Wang, D, Martinez Fiesco, J.A, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
7FXH
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2R)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, i.e. SMILES c1(c(c(c2c(n1)CCCCC2)c1ccccc1)C1=NN=NN1)[C@H]1CCCO1 with IC50=0.0496937 microM | | Descriptor: | (3P)-2-[(2R)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7FWY
 
 | | Crystal Structure of human FABP4 in complex with 2-[(3-chloro-5,6-dihydrobenzo[b][1]benzothiepin-6-yl)sulfanyl]acetic acid, i.e. SMILES c12c(Sc3c(C[C@H]1SCC(=O)O)cc(cc3)Cl)cccc2 with IC50=1.7 microM | | Descriptor: | Fatty acid-binding protein, adipocyte, SULFATE ION, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Boehringer, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6WXZ
 
 | | CRYSTAL STRUCTURE OF MYELOPEROXIDASE SUBFORM C (MPO) COMPLEX WITH Compound-29 A.K.A 7-(1,2-DIPHENYLETHYL)-1H-[1,2,3]TRIAZOLO[4,5-B]PYRIDIN-5-AMINE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(1R)-1,2-diphenylethyl]-3H-[1,2,3]triazolo[4,5-b]pyridin-5-amine, CALCIUM ION, ... | | Authors: | Khan, J.A. | | Deposit date: | 2020-05-12 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery and structure activity relationships of 7-benzyl triazolopyridines as stable, selective, and reversible inhibitors of myeloperoxidase.
Bioorg.Med.Chem., 28, 2020
|
|
7FXN
 
 | | Crystal Structure of human FABP4 in complex with 2-[(2S)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, i.e. SMILES c1(c(c(c2c(n1)CCCCC2)c1ccccc1)C1=NN=NN1)[C@@H]1CCCO1 with IC50=0.0357232 microM | | Descriptor: | (3P)-2-[(2S)-oxolan-2-yl]-4-phenyl-3-(1H-tetrazol-5-yl)-6,7,8,9-tetrahydro-5H-cyclohepta[b]pyridine, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Obst-Sander, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6BKU
 
 | | Crystal Structure of the Human CAMKK2B bound to GSK650394 | | Descriptor: | 2-cyclopentyl-4-(5-phenyl-1H-pyrrolo[2,3-b]pyridin-3-yl)benzoic acid, Calcium/calmodulin-dependent protein kinase kinase 2, FORMIC ACID | | Authors: | Counago, R.M, dos Reis, C.V, de Souza, G.P, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Human CAMKK2B bound to GSK650394
To Be Published
|
|
3KKV
 
 | | Structure of PKA with a protein Kinase B-selective inhibitor. | | Descriptor: | (2S)-1-{[6-furan-3-yl-5-(3-methyl-2H-indazol-5-yl)pyridin-3-yl]oxy}-3-(1H-indol-3-yl)propan-2-amine, PKI kinase inhibitor, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Elkins, P.A, Concha, N.O. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of PKA with a protein Kinase B-selective inhibitor.
To be Published
|
|
6P7G
 
 | | The co-crystal structure of BRAF(V600E) with PHI1 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-[(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-[4-({[2-(morpholin-4-yl)ethyl]amino}methyl)-3-(trifluoromethyl)phenyl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Agianian, B, Gavathiotis, E. | | Deposit date: | 2019-06-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibitors of BRAF dimers using an allosteric site.
Nat Commun, 11, 2020
|
|
6CDT
 
 | |
7P4F
 
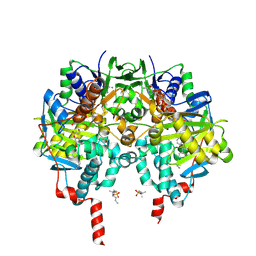 | | Crystal Structure of Monoamine Oxidase B in complex with inhibitor 1 | | Descriptor: | 4-(hydroxymethyl)-7-[[4-[[methyl-(phenylmethyl)amino]methyl]phenyl]methoxy]chromen-2-one, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Binda, C, Pisani, L. | | Deposit date: | 2021-07-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dual Reversible Coumarin Inhibitors Mutually Bound to Monoamine Oxidase B and Acetylcholinesterase Crystal Structures.
Acs Med.Chem.Lett., 13, 2022
|
|
7P4H
 
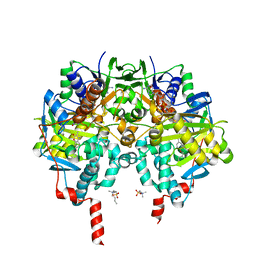 | | Crystal Structure of Monoamine Oxidase B in complex with inhibitor (+)-2 | | Descriptor: | 3,4-dimethyl-7-[[(3~{S})-piperidin-3-yl]methoxy]chromen-2-one, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Iacovino, L.G, Binda, C, Pisani, L. | | Deposit date: | 2021-07-11 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Reversible Coumarin Inhibitors Mutually Bound to Monoamine Oxidase B and Acetylcholinesterase Crystal Structures.
Acs Med.Chem.Lett., 13, 2022
|
|
6YXL
 
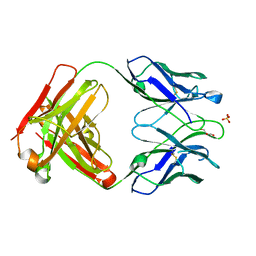 | | Crystal structure of ACPA F3 | | Descriptor: | ACPA F3 Fab fragment - heavy chain, ACPA F3 Fab fragment - light chain, GLYCEROL, ... | | Authors: | Ge, C, Holmdahl, R. | | Deposit date: | 2020-05-03 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Surface Ig variable domain glycosylation affects autoantigen binding and acts as threshold for human autoreactive B cell activation.
Sci Adv, 8, 2022
|
|
7RML
 
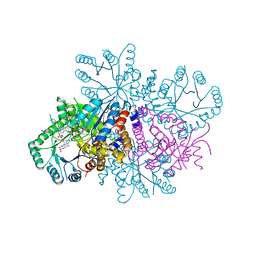 | | Neisseria meningitidis Methylenetetrahydrofolate reductase in complex with FAD | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pederick, J.L, Wegener, K.L, Salaemae, W, Bruning, J.B. | | Deposit date: | 2021-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of meningococcal methylenetetrahydrofolate reductase.
Protein Sci., 32, 2023
|
|
7H26
 
 | | PanDDA analysis group deposition -- Crystal Structure of ZIKV NS2B-NS3 protease in complex with Z1198317053 | | Descriptor: | (5-methyl-1H-pyrazolo[3,4-b]pyridin-1-yl)acetic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Ni, X, Godoy, A.S, Marples, P.G, Fairhead, M, Balcomb, B.H, Tomlinson, C.W.E, Koekemoer, L, Aschenbrenner, J.C, Lithgo, R.M, Thompson, W, Wild, C, Williams, E.P, Winokan, M, Chandran, A.V, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | PanDDA analysis group deposition of ZIKV NS2B-NS3 protease
To Be Published
|
|
7GYZ
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000035-001 | | Descriptor: | N-(4-methyl-3,4-dihydro-2H-1,4-benzoxazin-5-yl)-N'-(1H-pyrazolo[3,4-b]pyridin-5-yl)urea, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
6CJW
 
 | | Crystal Structure of Mnk2-D228G in Complex With Inhibitor | | Descriptor: | 3-(pyridin-4-yl)imidazo[1,2-b]pyridazine, MAP kinase-interacting serine/threonine-protein kinase 2, ZINC ION | | Authors: | Han, Q. | | Deposit date: | 2018-02-27 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structure-based Design of Pyridone-Aminal eFT508 Targeting Dysregulated Translation by Selective Mitogen-activated Protein Kinase Interacting Kinases 1 and 2 (MNK1/2) Inhibition.
J. Med. Chem., 61, 2018
|
|
7GZ4
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000018-001 | | Descriptor: | 5-[(2-fluorophenyl)sulfamoyl]-2-methyl-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7H09
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0011177-001 | | Descriptor: | (4M)-1-methyl-4-(4-{[(1R)-2-methyl-1-(3-oxo-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazin-6-yl)propyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-1H-pyrazole-5-carbonitrile, Papain-like protease nsp3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
7GZ1
 
 | | Crystal structure of SARS-CoV-2 NSP3 Macrodomain in complex with ASAP-0000061-001 | | Descriptor: | 3-chloro-N-(1H-pyrazolo[3,4-b]pyridin-5-yl)pyridine-4-carboxamide, Non-structural protein 3 | | Authors: | Aschenbrenner, J.C, Fearon, D, Tomlinson, C.W.E, Marples, P.G, Fairhead, M, Balcomb, B.H, Chandran, A.V, Godoy, A.S, Koekemoer, L, Lithgo, R.M, Ni, X, Thompson, W, Wang, S, Wild, C, Williams, E.P, Winokan, M, Walsh, M.A, von Delft, F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Group deposition of SARS-CoV-2 NSP3 Macrodomain in complex with inhibitors from the ASAP AViDD centre
To Be Published
|
|
5E61
 
 | |
