5TCS
 
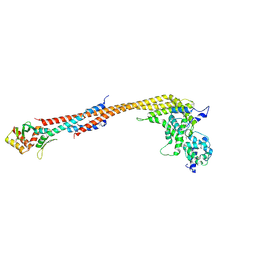 | | Crystal structure of a Dwarf Ndc80 Tetramer | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Harrison, S.C. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-23 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8313 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
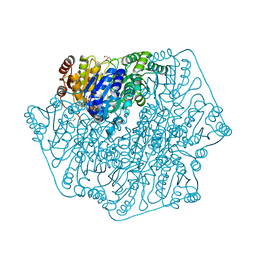
4MPR
 
 | |
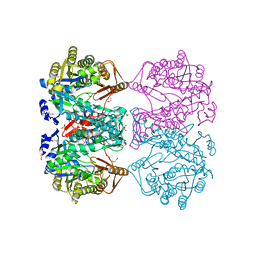
3DO6
 
 | |
5KID
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | CHLORIDE ION, GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
5KHP
 
 | | Tightening the Recognition of Tetravalent Zr and Th Complexes by the Siderophore-Binding Mammalian Protein Siderocalin for Theranostic Applications | | Descriptor: | GLYCEROL, N,N'-(butane-1,4-diyl)bis(N-{3-[(2,3-dihydroxybenzene-1-carbonyl)amino]propyl}-2,3-dihydroxybenzamide), Neutrophil gelatinase-associated lipocalin, ... | | Authors: | Rupert, P.B, Strong, R.K. | | Deposit date: | 2016-06-15 | | Release date: | 2017-04-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered Recognition of Tetravalent Zirconium and Thorium by Chelator-Protein Systems: Toward Flexible Radiotherapy and Imaging Platforms.
Inorg Chem, 55, 2016
|
|
4PXT
 
 | |
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
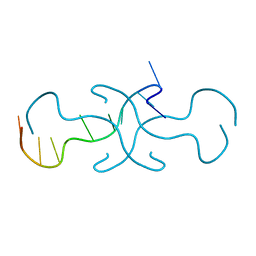
4RIP
 
 | |
3ALB
 
 | | Cyclic Lys48-linked tetraubiquitin | | Descriptor: | SULFATE ION, ubiquitin | | Authors: | Satoh, T, Sakata, E, Yamamoto, S, Yamaguchi, Y, Sumiyoshi, A, Wakatsuki, S, Kato, K. | | Deposit date: | 2010-07-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of cyclic Lys48-linked tetraubiquitin
Biochem.Biophys.Res.Commun., 2010
|
|
3CRP
 
 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, SODIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-04-07 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
3GSI
 
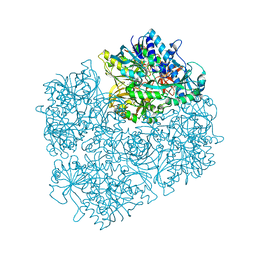 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
105D
 
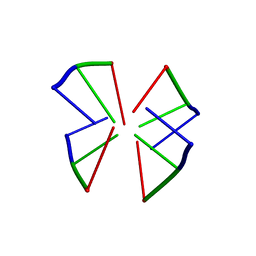 | |
106D
 
 | |
4CQL
 
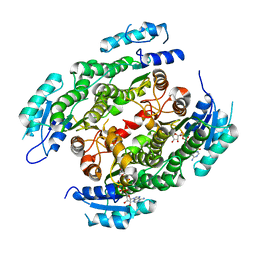 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD | | Descriptor: | CARBONYL REDUCTASE FAMILY MEMBER 4, ESTRADIOL 17-BETA-DEHYDROGENASE 8, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Venkatesan, R, Sah-Teli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastaniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
3CK4
 
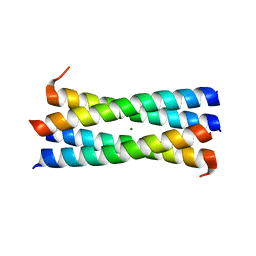 | | A heterospecific leucine zipper tetramer | | Descriptor: | GCN4 leucine zipper, MAGNESIUM ION | | Authors: | Liu, J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A heterospecific leucine zipper tetramer.
Chem.Biol., 15, 2008
|
|
4D2H
 
 | |
4RIM
 
 | |
4MPW
 
 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzofuran-3,5-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | ACETATE ION, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel, Nonpeptidic, Orally Active Bivalent Inhibitor of Human beta-Tryptase.
Pharmacology, 102, 2018
|
|
3NWH
 
 | | Crystal structure of BST2/Tetherin | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Schubert, H.L, Zhai, Q, Hill, C.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7CD4
 
 | | Crystal structure of the S103F mutant of Bacillus subtilis (natto) YabJ protein. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fujimoto, Z, Kishine, N, Kimura, K. | | Deposit date: | 2020-06-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tetramer formation of Bacillus subtilis YabJ protein that belongs to YjgF/YER057c/UK114 family.
Biosci.Biotechnol.Biochem., 85, 2021
|
|
7CD3
 
 | |
7CD2
 
 | |
5A3F
 
 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
3NV7
 
 | |
4CQM
 
 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD and NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBONYL REDUCTASE FAMILY MEMBER 4, ... | | Authors: | Venkatesan, R, SahTeli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastoniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
