6QCI
 
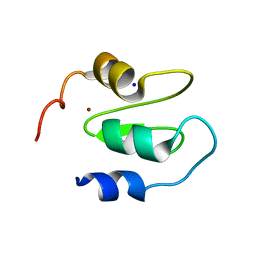 | | Structure of XIAP-BIR1 V86E mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase XIAP, SODIUM ION, ... | | Authors: | Sorrentino, L, Cossu, F, Milani, M, Mastrangelo, E. | | Deposit date: | 2018-12-28 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Activity Relationship of NF023 Derivatives Binding to XIAP-BIR1.
Chemistryopen, 8, 2019
|
|
3FUQ
 
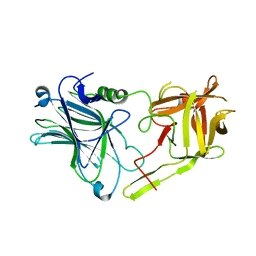 | | Glycosylated SV2 and Gangliosides as Dual Receptors for Botulinum Neurotoxin Serotype F | | Descriptor: | BoNT/F (Neurotoxin type F) | | Authors: | Fu, Z, Chen, C, Barbieri, J.T, Kim, J.-J.P, Baldwin, M.R. | | Deposit date: | 2009-01-14 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glycosylated SV2 and gangliosides as dual receptors for botulinum neurotoxin serotype F
Biochemistry, 48, 2009
|
|
6QE9
 
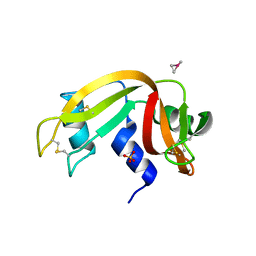 | | The X-ray structure of the adduct formed in the reaction between bovine pancreatic ribonuclease and complex I, a pentacoordinate Pt(II) compound containing 2,9-dimethyl-1,10-phenanthroline, dimethylfumarate, methyl and iodine as ligands | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, pentacoordinate Pt(II) compound | | Authors: | Merlino, A, Ferraro, G. | | Deposit date: | 2019-01-07 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Reaction with Proteins of a Five-Coordinate Platinum(II) Compound.
Int J Mol Sci, 20, 2019
|
|
6Q2Y
 
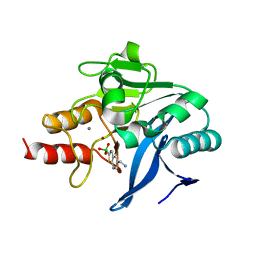 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd3 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, Metallo-beta-lactamase type 2, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-12-03 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6RYV
 
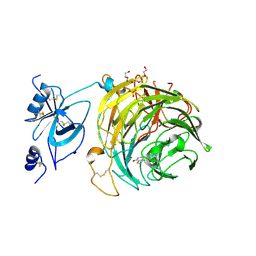 | | Copper oxidase from Colletotrichum graminicola | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Kelch domain-containing protein, ... | | Authors: | Offen, W.A, Henrissat, B, Davies, G.J. | | Deposit date: | 2019-06-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Acs Catalysis, 2020
|
|
3JXI
 
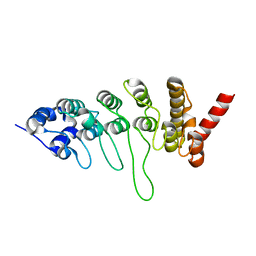 | |
6WC5
 
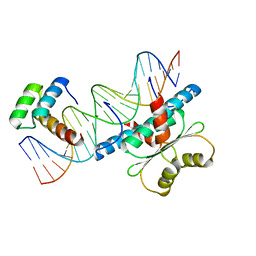 | | Crystal Structure of a Ternary MEF2B/NKX2-5/myocardin enhancer DNA Complex | | Descriptor: | Homeobox protein Nkx-2.5, Myocardin enhancer DNA, Myocyte-specific enhancer factor 2B | | Authors: | Chen, L, Lei, X. | | Deposit date: | 2020-03-29 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of Ternary Complexes of MEF2 and NKX2-5 Bound to DNA Reveal a Disease Related Protein-Protein Interaction Interface.
J.Mol.Biol., 432, 2020
|
|
6RZA
 
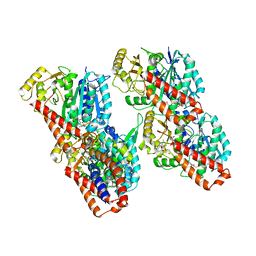 | | Cryo-EM structure of the human inner arm dynein DNAH7 microtubule binding domain bound to microtubules | | Descriptor: | Cytoplasmic dynein 1 heavy chain 1,Dynein heavy chain 7, axonemal,Cytoplasmic dynein 1 heavy chain 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lacey, S.E, He, S, Scheres, S.H.W, Carter, A.P. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM of dynein microtubule-binding domains shows how an axonemal dynein distorts the microtubule.
Elife, 8, 2019
|
|
6S04
 
 | | Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid | | Descriptor: | ACETATE ION, GLYCEROL, N-glycolyl-beta-neuraminic acid, ... | | Authors: | Bule, P, Blagova, E, Chuzel, L, Taron, C.H, Davies, G.J. | | Deposit date: | 2019-06-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inverting family GH156 sialidases define an unusual catalytic motif for glycosidase action.
Nat Commun, 10, 2019
|
|
6QG0
 
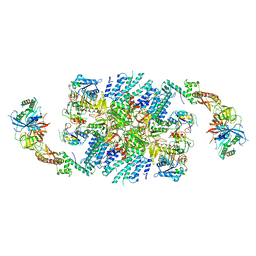 | | Structure of eIF2B-eIF2 (phosphorylated at Ser51) complex (model 1) | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Llacer, J.L, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for the inhibition of translation through eIF2 alpha phosphorylation.
Nat Commun, 10, 2019
|
|
3JZQ
 
 | |
6Q45
 
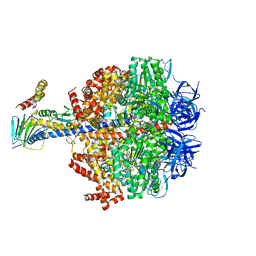 | | F1-ATPase from Fusobacterium nucleatum | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Petri, J, Nakatani, Y, Montgomery, M.G, Ferguson, S.A, Aragao, D, Leslie, A.G.W, Heikal, A, Walker, J.E, Cook, G.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of F1-ATPase from the obligate anaerobe Fusobacterium nucleatum.
Open Biology, 9, 2019
|
|
6RGY
 
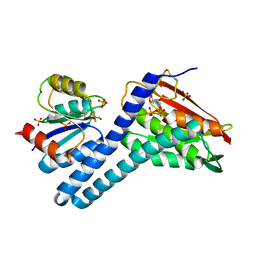 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RI2
 
 | | Single crystal serial study of the inhibition of laccases from Steccherinum murashkinskyi by chloride anions at sub-atomic resolution. Twentieth structure of the series with 4065 KGy dose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Polyakov, K.M, Gavryushov, S, Fedorova, T.V, Glazunova, O.A, Popov, A.N. | | Deposit date: | 2019-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | The subatomic resolution study of laccase inhibition by chloride and fluoride anions using single-crystal serial crystallography: insights into the enzymatic reaction mechanism.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QKT
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - Tyr219Phe - Fluoroacetate soaked 24hr - Glycolate bound | | Descriptor: | Fluoroacetate dehalogenase, GLYCOLIC ACID | | Authors: | Mehrabi, P, Kim, T.H, Prosser, R.S, Pai, E.F. | | Deposit date: | 2019-01-30 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.512 Å) | | Cite: | Substrate-Based Allosteric Regulation of a Homodimeric Enzyme.
J.Am.Chem.Soc., 141, 2019
|
|
6S6Z
 
 | | Structure of beta-Galactosidase from Thermotoga maritima | | Descriptor: | Beta-galactosidase, MAGNESIUM ION | | Authors: | Miguez-Amil, S, Jimenez-Ortega, E, Ramirez Escudero, M, Sanz-Aparicio, J, Fernandez-Leiro, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | The cryo-EM Structure ofThermotoga maritimabeta-Galactosidase: Quaternary Structure Guides Protein Engineering.
Acs Chem.Biol., 15, 2020
|
|
6WIS
 
 | | Copper resistance protein copG- Form 1 | | Descriptor: | ACETATE ION, COPPER (II) ION, DUF411 domain-containing protein, ... | | Authors: | Hausrath, A.C, Ly, A, McEvoy, M.M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-06-24 | | Last modified: | 2020-08-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The bacterial copper resistance protein CopG contains a cysteine-bridged tetranuclear copper cluster.
J.Biol.Chem., 295, 2020
|
|
6Q50
 
 | | Structure of MPT-4, a mannose phosphorylase from Leishmania mexicana, in complex with phosphate ion | | Descriptor: | 1,2-ETHANEDIOL, MPT-4, PHOSPHATE ION, ... | | Authors: | Sobala, L.F, Males, A, Bastidas, L.M, Ward, T, Sernee, M.F, Ralton, J.E, Nero, T.L, Cobbold, S, Kloehn, J, Viera-Lara, M, Stanton, L, Hanssen, E, Parker, M.W, Williams, S.J, McConville, M.J, Davies, G.J. | | Deposit date: | 2018-12-06 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Family of Dual-Activity Glycosyltransferase-Phosphorylases Mediates Mannogen Turnover and Virulence in Leishmania Parasites.
Cell Host Microbe, 26, 2019
|
|
3K42
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-dependent mannose-6-phosphate receptor, SN-GLYCEROL-1-PHOSPHATE, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
6Q6C
 
 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | 1,2-ETHANEDIOL, Kunitz-type conkunitzin-S1, NITRATE ION, ... | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6WGY
 
 | |
6RYX
 
 | | Copper oxidase from Colletotrichum graminicola | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Offen, W.A, Henrissat, B, Davies, G.J. | | Deposit date: | 2019-06-12 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of a Fungal Copper Radical Oxidase with High Catalytic Efficiency toward 5-Hydroxymethylfurfural and Benzyl Alcohols for Bioprocessing
Acs Catalysis, 2020
|
|
3K77
 
 | | X-ray crystal structure of XRCC1 | | Descriptor: | DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, London, R.E. | | Deposit date: | 2009-10-12 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6WDV
 
 | |
6QIH
 
 | |
