7W9T
 
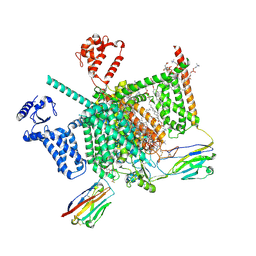 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
4C22
 
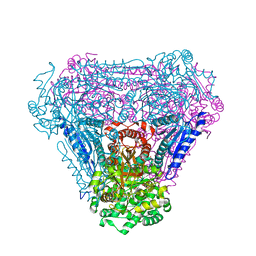 | | L-Fucose Isomerase In Complex With Fuculose | | Descriptor: | 1,2-ETHANEDIOL, L-FUCOSE ISOMERASE, L-Fuculose open form, ... | | Authors: | Higgins, M.A, Suits, M.D.L, Marsters, C, Boraston, A.B. | | Deposit date: | 2013-08-16 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Functional Analysis of Fucose-Processing Enzymes from Streptococcus Pneumoniae.
J.Mol.Biol., 426, 2014
|
|
5W5Q
 
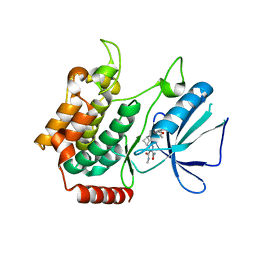 | | MAP4K4 in complex with inhibitor compound 12 (N3-methyl-10-(3-methyl-3-(5-methyloxazol-2-yl)but-1-yn-1-yl)-6,7-dihydro-5H-5,7-methanobenzo[c]imidazo[1,2-a]azepine-2,3-dicarboxamide) | | Descriptor: | (5s,7s)-N~3~-methyl-10-[3-methyl-3-(5-methyl-1,3-oxazol-2-yl)but-1-yn-1-yl]-6,7-dihydro-5H-5,7-methanoimidazo[2,1-a][2]benzazepine-2,3-dicarboxamide, Mitogen-activated protein kinase kinase kinase kinase 4 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2017-06-15 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure Based Design of Potent Selective Inhibitors of Protein Kinase D1 (PKD1).
Acs Med.Chem.Lett., 10, 2019
|
|
1B43
 
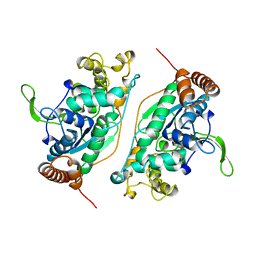 | | FEN-1 FROM P. FURIOSUS | | Descriptor: | PROTEIN (FEN-1) | | Authors: | Hosfield, D.J, Mol, C.D, Shen, B, Tainer, J.A. | | Deposit date: | 1999-01-05 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity.
Cell(Cambridge,Mass.), 95, 1998
|
|
5DXU
 
 | | p110delta/p85alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
1T1V
 
 | | Crystal Structure of the Glutaredoxin-like Protein SH3BGRL3 at 1.6 A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, SH3 domain-binding glutamic acid-rich protein-like 3, ... | | Authors: | Nardini, M, Mazzocco, M, Massaro, M, Maffei, M, Vergano, A, Donadini, A, Scartezzini, M, Bolognesi, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the glutaredoxin-like protein SH3BGRL3 at 1.6 A resolution
Biochem.Biophys.Res.Commun., 318, 2004
|
|
5DXH
 
 | | p110alpha/p85alpha with compound 5 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, methyl {2-[4-(2-chlorophenyl)-4H-1,2,4-triazol-3-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepin-8-yl}carbamate | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
2YHW
 
 | | High-resolution crystal structures of N-Acetylmannosamine kinase: Insights about substrate specificity, activity and inhibitor modelling. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ACETATE ION, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Zimmer, R, Tauberger, E, Reutter, W, Saenger, W, Fan, H, Moniot, S. | | Deposit date: | 2011-05-08 | | Release date: | 2012-02-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
4ZZT
 
 | | Geotrichum candidum Cel7A structure complex with thio-linked cellotriose at 1.56A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, GLYCEROL, ... | | Authors: | Borisova, A.S, Stahlberg, J. | | Deposit date: | 2015-04-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Sequencing, Biochemical Characterization, Crystal Structure and Molecular Dynamics of Cellobiohydrolase Cel7A from Geotrichum Candidum 3C.
FEBS J., 282, 2015
|
|
5A82
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 4-(3R,4R) -4-(3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)aminopiperidin-3- yloxymethyl)-1-thiane-1,1-dione | | Descriptor: | 1,2-ETHANEDIOL, 8-[[(3R,4R)-3-[[1,1-bis(oxidanylidene)thian-4-yl]methoxy]piperidin-4-yl]amino]-3-methyl-1H-1,7-naphthyridin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-07-11 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Based Optimization of Naphthyridones Into Potent Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
3I2E
 
 | | Crystal structure of human dimethylarginine dymethylaminohydrolase-1 (DDAH-1) | | Descriptor: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | Authors: | Monzingo, A.F, Wang, Y, Hu, S, Schaller, T.H, Robertus, J.D, Fast, W. | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Developing dual and specific inhibitors of dimethylarginine dimethylaminohydrolase-1 and nitric oxide synthase: toward a targeted polypharmacology to control nitric oxide.
Biochemistry, 48, 2009
|
|
1GM9
 
 | | Crystal structures of penicillin acylase enzyme-substrate complexes: Structural insights into the catalytic mechanism | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-[(2S,4S,6R)-2-(DIHYDROXYMETHYL)-4-HYDROXY-3,3-DIMETHYL-7-OXO-4LAMBDA~4~-THIA-1-AZABICYCLO[3.2.0]HEPT-6-YL]-2-PHENYLAC ETAMIDE, ... | | Authors: | McVey, C.E, Walsh, M.A, Dodson, G.G, Wilson, K.S, Brannigan, J.A. | | Deposit date: | 2001-09-12 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Penicillin Acylase Enzyme- Substrate Complexes: Structural Insights Into the Catalytic Mechanism
J.Mol.Biol., 313, 2001
|
|
5I8B
 
 | | CBP in complex with Cpd23 ((R)-6-(3-(benzyloxy)phenyl)-4-methyl-1,3,4,5-tetrahydro-2H-benzo[b][1,4]diazepin-2-one) | | Descriptor: | (4R)-6-[3-(benzyloxy)phenyl]-4-methyl-1,3,4,5-tetrahydro-2H-1,5-benzodiazepin-2-one, 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5218 Å) | | Cite: | Fragment-Based Discovery of a Selective and Cell-Active Benzodiazepinone CBP/EP300 Bromodomain Inhibitor (CPI-637).
Acs Med.Chem.Lett., 7, 2016
|
|
5TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1: ORTHORHOMBIC P222(1) CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
1UL7
 
 | | Solution structure of kinase associated domain 1 of mouse MAP/microtubule affinity-regulating kinase 3 | | Descriptor: | MAP/microtubule affinity-regulating kinase 3 | | Authors: | Tochio, N, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-10 | | Release date: | 2004-03-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kinase-associated domain 1 of mouse microtubule-associated protein/microtubule affinity-regulating kinase 3
Protein Sci., 15, 2006
|
|
2X7A
 
 | | Structural basis of HIV-1 tethering to membranes by the Bst2-tetherin ectodomain | | Descriptor: | BONE MARROW STROMAL ANTIGEN 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Natrajan, G, McCarthy, A.A, Weissenhorn, W. | | Deposit date: | 2010-02-25 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural Basis of HIV-1 Tethering to Membranes by the Bst-2/Tetherin Ectodomain.
Cell Host Microbe, 7, 2010
|
|
6UUH
 
 | |
5TTS
 
 | | Jak3 with covalent inhibitor 4 | | Descriptor: | 1-{(3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
1HX9
 
 | | CRYSTAL STRUCTURE OF TEAS W273S FORM 1 | | Descriptor: | 1-HYDROXY-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENE PHOSPHONIC ACID, 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.S, Rising, K.A, Chappell, J, Noel, J.P. | | Deposit date: | 2001-01-12 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Single Active Site Mutations Change the Specificity of a Sesquiterpene Cyclase
To be Published
|
|
1V82
 
 | | Crystal structure of human GlcAT-P apo form | | Descriptor: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | Authors: | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | Deposit date: | 2003-12-27 | | Release date: | 2004-05-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
2P94
 
 | |
4GW3
 
 | | Crystal Structure of the Lipase from Proteus mirabilis | | Descriptor: | CALCIUM ION, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Korman, T.P. | | Deposit date: | 2012-08-31 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Proteus mirabilis Lipase, a Novel Lipase from the Proteus/Psychrophilic Subfamily of Lipase Family I.1.
Plos One, 7, 2012
|
|
2P93
 
 | |
3N8U
 
 | |
5UUK
 
 | |
