3FQO
 
 | | Staphylococcus aureus F98Y mutant dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(2,5-dimethoxyphenyl)prop-1-ynyl]-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
2OIZ
 
 | |
2O19
 
 | | Structure of E. coli topoisomersae III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 5.5 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, DiGate, R, Mondragon, A. | | Deposit date: | 2006-11-28 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
3FST
 
 | |
2O3A
 
 | | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus | | Descriptor: | UPF0106 protein AF_0751 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Wu, B, Slocombe, A, Sridhar, V, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-12-01 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a protein AF_0751 from Archaeoglobus fulgidus
To be Published
|
|
3G59
 
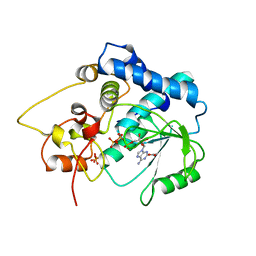 | |
3POW
 
 | |
2NSN
 
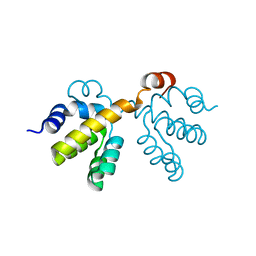 | |
3G6K
 
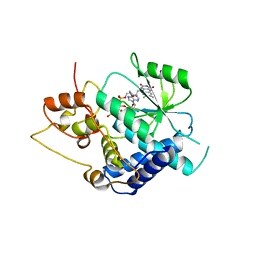 | | Crystal Structure of Candida glabrata FMN Adenylyltransferase in complex with FAD and Inorganic Pyrophosphate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FMN adenylyltransferase, MAGNESIUM ION, ... | | Authors: | Huerta, C, Machius, M, Zhang, H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure and mechanism of a eukaryotic FMN adenylyltransferase.
J.Mol.Biol., 389, 2009
|
|
3PPW
 
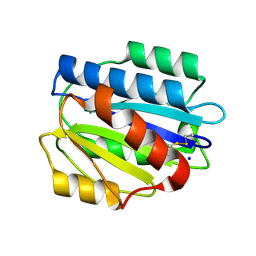 | | Crystal structure of the D1596A mutant of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | SODIUM ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
2NW3
 
 | | Crystal structure of HLA-B*3508 presenting EBV peptide EPLPQGQLTAY at 1.7A | | Descriptor: | Beta-2-microglobulin, EBV peptide EPLPQGQLTAY, HLA class I histocompatibility antigen, ... | | Authors: | Tynan, F.E, Reid, H.H, Rossjohn, J. | | Deposit date: | 2006-11-14 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule
Nat.Immunol., 8, 2007
|
|
3FXL
 
 | | Crystal Structure of Human Protein phosphatase 1A (PPM1A) Bound with Citrate at 1 mM of Mn2+ | | Descriptor: | CITRATE ANION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Hu, T, Wang, L, Wang, K, Jiang, H, Shen, X. | | Deposit date: | 2009-01-21 | | Release date: | 2010-01-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the Mn2+-dependent activation of human PPM1A
To be published
|
|
3FQD
 
 | | Crystal Structure of the S. pombe Rat1-Rai1 Complex | | Descriptor: | 5'-3' exoribonuclease 2, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the 5'-->3' exoribonuclease Rat1 and its activating partner Rai1.
Nature, 458, 2009
|
|
3PT5
 
 | | Crystal structure of NanS | | Descriptor: | NANS (YJHS), A 9-O-acetyl N-acetylneuraminic acid esterase | | Authors: | Ruane, K.M, Rangarajan, E.S, Proteau, A, Schrag, J.D, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-12-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7.
Protein Sci., 20, 2011
|
|
3PU1
 
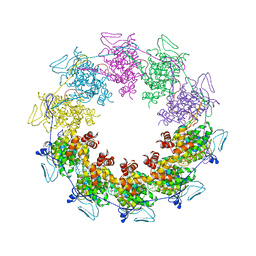 | |
2NXV
 
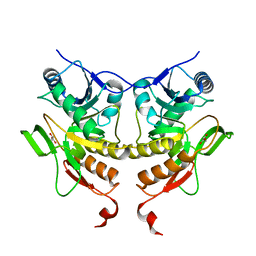 | |
3PV4
 
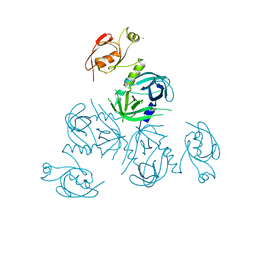 | | Structure of Legionella fallonii DegQ (Delta-PDZ2 variant) | | Descriptor: | CADMIUM ION, DegQ | | Authors: | Wrase, R, Scott, H, Hilgenfeld, R, Hansen, G. | | Deposit date: | 2010-12-06 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Legionella HtrA homologue DegQ is a self-compartmentizing protease that forms large 12-meric assemblies.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2NY5
 
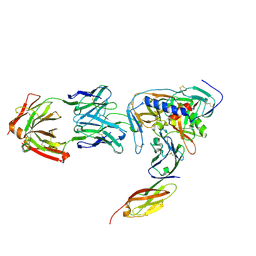 | | HIV-1 gp120 Envelope Glycoprotein (M95W, W96C, I109C, T257S, V275C, S334A, S375W, Q428C, A433M) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
3FQZ
 
 | | Staphylococcus aureus dihydrofolate reductase complexed with NADPH and 2,4-diamino-5-[3-(3-methoxy-4-phenylphenyl)but-1-ynyl]-6-methylpyrimidine | | Descriptor: | 5-[(3S)-3-(2-methoxybiphenyl-4-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Trimethoprim-sensitive dihydrofolate reductase | | Authors: | Anderson, A.C, Frey, K.M, Liu, J, Lombardo, M.N. | | Deposit date: | 2009-01-07 | | Release date: | 2009-03-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of wild-type and mutant methicillin-resistant Staphylococcus aureus dihydrofolate reductase reveal an alternate conformation of NADPH that may be linked to trimethoprim resistance.
J.Mol.Biol., 387, 2009
|
|
3PWE
 
 | | Crystal structure of the E. coli beta clamp mutant R103C, I305C, C260S, C333S at 2.2A resolution | | Descriptor: | DNA polymerase III subunit beta | | Authors: | Marzahn, M.R, Robbins, A.H, McKenna, R, Bloom, L.B. | | Deposit date: | 2010-12-08 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | The E. coli clamp loader can actively pry open the beta-sliding clamp
J.Biol.Chem., 286, 2011
|
|
3FWK
 
 | |
3PXH
 
 | | Tandem Ig domains of tyrosine phosphatase LAR | | Descriptor: | Receptor-type tyrosine-protein phosphatase F, SULFATE ION | | Authors: | Biersmith, B.H, Bouyain, S. | | Deposit date: | 2010-12-09 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0009 Å) | | Cite: | The Immunoglobulin-like Domains 1 and 2 of the Protein Tyrosine Phosphatase LAR Adopt an Unusual Horseshoe-like Conformation.
J.Mol.Biol., 408, 2011
|
|
3UTQ
 
 | | Human HLA-A*0201-ALWGPDPAAA | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Sewell, A.K, Bulek, A.M. | | Deposit date: | 2011-11-26 | | Release date: | 2012-01-25 | | Last modified: | 2012-03-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes.
Nat.Immunol., 13, 2012
|
|
3UC9
 
 | |
3G0G
 
 | | Crystal structure of dipeptidyl peptidase IV in complex with a pyrimidinone inhibitor 3 | | Descriptor: | 2-({2-[(3R)-3-aminopiperidin-1-yl]-5-bromo-6-oxopyrimidin-1(6H)-yl}methyl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, Z, Wallace, M.B, Feng, J, Stafford, J.A, Kaldor, S.W, Shi, L, Skene, R.J, Aertgeerts, K, Lee, B, Jennings, A, Xu, R, Kassel, D, Webb, D.R, Gwaltney, S.L. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Design and Synthesis of Pyrimidinone and Pyrimidinedione Inhibitors of Dipeptidyl Peptidase IV.
J.Med.Chem., 54, 2011
|
|
