5V80
 
 | |
2QED
 
 | | Crystal structure of Salmonella thyphimurium LT2 glyoxalase II | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Hydroxyacylglutathione hydrolase | | Authors: | Leite, N.R, Campos Bermudez, V.A, Krogh, R, Oliva, G, Soncini, F.C, Vila, A.J. | | Deposit date: | 2007-06-25 | | Release date: | 2007-10-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Biochemical and Structural Characterization of Salmonella typhimurium Glyoxalase II: New Insights into Metal Ion Selectivity
Biochemistry, 46, 2007
|
|
3DN1
 
 | | Chloropentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1-chloro-2,3,4,5,6-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
1TLD
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-TRYPSIN AT 1.5 ANGSTROMS RESOLUTION IN A CRYSTAL FORM WITH LOW MOLECULAR PACKING DENSITY. ACTIVE SITE GEOMETRY, ION PAIRS AND SOLVENT STRUCTURE | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, SULFATE ION | | Authors: | Bartunik, H.D, Summers, L.J, Bartsch, H.H. | | Deposit date: | 1989-07-24 | | Release date: | 1990-01-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bovine beta-trypsin at 1.5 A resolution in a crystal form with low molecular packing density. Active site geometry, ion pairs and solvent structure.
J.Mol.Biol., 210, 1989
|
|
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
1RNB
 
 | |
4COQ
 
 | | The complex of alpha-Carbonic anhydrase from Thermovibrio ammonificans with inhibitor sulfanilamide. | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CARBONATE DEHYDRATASE, CHLORIDE ION, ... | | Authors: | James, P, Isupov, M.N, Sayer, C, Berg, S, Lioliou, M, Kotlar, H, Littlechild, J.A. | | Deposit date: | 2014-01-30 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Structure of a Tetrameric [Alpha]-Carbonic Anhydrase from Thermovibrio Ammonificans Reveals a Core Formed Around Intermolecular Disulfides that Contribute to its Thermostability
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2RLG
 
 | |
3ZUU
 
 | | The structure of OST1 (D160A, S175D) kinase in complex with gold | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, Serine/threonine-protein kinase SRK2E | | Authors: | Yunta, C, Martinez-Ripoll, M, Albert, A. | | Deposit date: | 2011-07-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress.
J. Mol. Biol., 414, 2011
|
|
2HBE
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (BETA CHAIN), N-BUTYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
2FVD
 
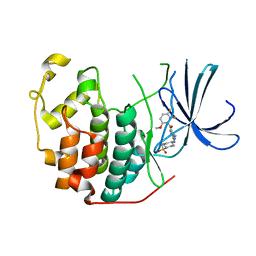 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
4DR8
 
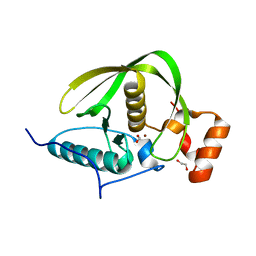 | | Crystal structure of a peptide deformylase from Synechococcus elongatus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Lorimer, D, Abendroth, J, Craig, T, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2012-02-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
4MCD
 
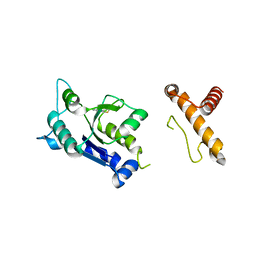 | | hinTrmD in complex with 5-PHENYLTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lahiri, S. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
2Z53
 
 | |
3G5S
 
 | | Crystal structure of Thermus thermophilus TrmFO in complex with glutathione | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, ... | | Authors: | Nishimasu, H, Ishitani, R, Hori, H, Nureki, O. | | Deposit date: | 2009-02-05 | | Release date: | 2009-05-19 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Atomic structure of a folate/FAD-dependent tRNA T54 methyltransferase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3Q92
 
 | | X-ray Structure of ketohexokinase in complex with a pyrimidopyrimidine analog 1 | | Descriptor: | Ketohexokinase, N~8~-(cyclopropylmethyl)-N~4~-[2-(methylsulfanyl)phenyl]-2-(piperazin-1-yl)pyrimido[5,4-d]pyrimidine-4,8-diamine, SULFATE ION | | Authors: | Abad, M.C. | | Deposit date: | 2011-01-07 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibitors of Ketohexokinase: Discovery of Pyrimidinopyrimidines with Specific Substitution that Complements the ATP-Binding Site.
ACS Med Chem Lett, 2, 2011
|
|
4DF3
 
 | |
4MCC
 
 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
2JUK
 
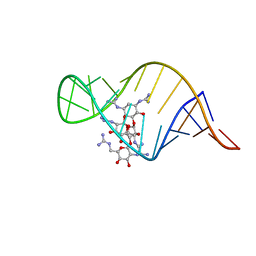 | | guanidino neomycin B recognition of an HIV-1 RNA helix | | Descriptor: | (1S,2R,3S,4R,6S)-4,6-bis{[amino(iminio)methyl]amino}-2-{[3-O-(2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranosyl)-beta-D-arabinofuranosyl]oxy}-3-hydroxycyclohexyl 2,6-bis{[amino(iminio)methyl]amino}-2,6-dideoxy-beta-L-glucopyranoside, HIV-1 frameshift site RNA | | Authors: | Staple, D.W, Venditti, V, Niccolai, N, Elson-Schwab, L, Tor, Y, Butcher, S.E. | | Deposit date: | 2007-08-30 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Guanidinoneomycin B Recognition of an HIV-1 RNA Helix.
Chembiochem, 9, 2008
|
|
3OHG
 
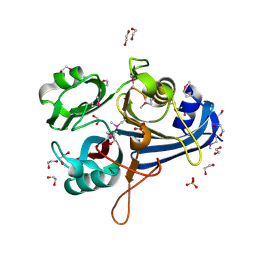 | |
2PYU
 
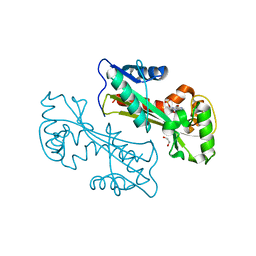 | | Structure of the E. coli inosine triphosphate pyrophosphatase RgdB in complex with IMP | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, Inosine Triphosphate Pyrophosphatase RdgB | | Authors: | Singer, A.U, Proudfoot, M, Skarina, T, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2007-05-16 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Molecular basis of the antimutagenic activity of the house-cleaning inosine triphosphate pyrophosphatase RdgB from Escherichia coli.
J.Mol.Biol., 374, 2007
|
|
2G47
 
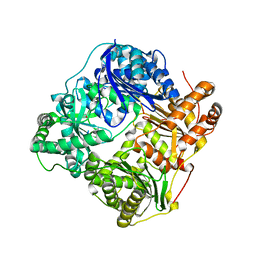 | |
1PJV
 
 | | Cobatoxin 1 from Centruroides noxius Scorpion venom: Chemical Synthesis, 3-D Structure in Solution, Pharmacology and Docking on K+ channels | | Descriptor: | Cobatoxin 1 | | Authors: | Mosbah, A, Jouirou, B, Visan, V, Grissmer, S, El Ayeb, M, Rochat, H, De Waard, M, Mabrouk, K, Sabatier, J.M. | | Deposit date: | 2003-06-03 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Cobatoxin 1 from Centruroides noxius scorpion venom: chemical synthesis, three-dimensional structure in solution, pharmacology and docking on K+ channels.
Biochem.J., 377, 2004
|
|
1PN4
 
 | | Crystal structure of 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 complexed with (3R)-hydroxydecanoyl-CoA. | | Descriptor: | 1,2-ETHANEDIOL, 3R-HYDROXYDECANOYL-COENZYME A, Peroxisomal hydratase-dehydrogenase-epimerase | | Authors: | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2003-06-12 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
3ZHI
 
 | | N-terminal domain of the CI repressor from bacteriophage TP901-1 | | Descriptor: | CI | | Authors: | Frandsen, K.H, Rasmussen, K.K, Poulsen, J.N, Lo Leggio, L. | | Deposit date: | 2012-12-21 | | Release date: | 2013-12-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding of the N-Terminal Domain of the Lactococcal Bacteriophage Tp901-1 Ci Repressor to its Target DNA: A Crystallography, Small Angle Scattering, and Nuclear Magnetic Resonance Study.
Biochemistry, 52, 2013
|
|
