7Z50
 
 | |
1ZMF
 
 | | C domain of human cyclophilin-33(hcyp33) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Wang, T, Yun, C.-H, Gu, S.-Y, Chang, W.-R, Liang, D.-C. | | Deposit date: | 2005-05-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88A crystal structure of the C domain of hCyP33: A novel domain of peptidyl-prolyl cis-trans isomerase
Biochem.Biophys.Res.Commun., 333, 2005
|
|
5C3T
 
 | | PD-1 binding domain from human PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Dubin, G, Holak, T.A. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Complex of Human Programmed Death 1, PD-1, and Its Ligand PD-L1.
Structure, 23, 2015
|
|
7YVZ
 
 | | Structure of Caenorhabditis elegans CISD-1/mitoNEET | | Descriptor: | CDGSH iron-sulfur domain-containing protein 1 (CISD-1/mitoNEET), FE2/S2 (INORGANIC) CLUSTER | | Authors: | Hasegawa, K, Hagiuda, E, Taguchi, A.T, Geldenhuys, W, Iwasaki, T, Kumasaka, T. | | Deposit date: | 2022-08-20 | | Release date: | 2022-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and biological evaluation of Caenorhabditis elegans CISD-1/mitoNEET, a KLP-17 tail domain homologue, supports attenuation of paraquat-induced oxidative stress through a p38 MAPK-mediated antioxidant defense response.
Adv Redox Res, 6, 2022
|
|
6N1E
 
 | | Crystal structure of X. citri phosphoglucomutase in complex with 1-methyl-glucose 6-phosphate | | Descriptor: | 1-deoxy-7-O-phosphono-alpha-D-gluco-hept-2-ulopyranose, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Beamer, L.J, Stiers, K.M. | | Deposit date: | 2018-11-08 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis, Derivatization, and Structural Analysis of Phosphorylated Mono-, Di-, and Trifluorinated d-Gluco-heptuloses by Glucokinase: Tunable Phosphoglucomutase Inhibition.
Acs Omega, 4, 2019
|
|
4JT8
 
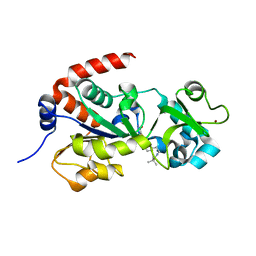 | | Crystal Structure of human SIRT3 with ELT inhibitor 28 [4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide[ | | Descriptor: | 4-(4-{2-[(2,2-dimethylpropanoyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ... | | Authors: | Dai, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Thieno[3,2-d]pyrimidine-6-carboxamides as Potent Inhibitors of SIRT1, SIRT2, and SIRT3.
J.Med.Chem., 56, 2013
|
|
4JTW
 
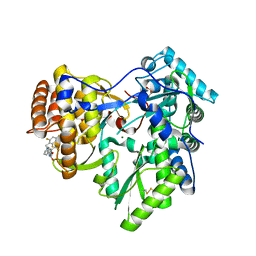 | | Crystal structure of HCV NS5B polymerase in complex with coupound 1 | | Descriptor: | 1-(2,4,6-trifluorobenzyl)-6-[2-(trifluoromethyl)phenoxy]quinazolin-4(1H)-one, GLYCEROL, Genome polyprotein, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-03-24 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Dynamics Simulations and Structure-Based Rational Design Lead to Allosteric HCV NS5B Polymerase Thumb Pocket 2 Inhibitor with Picomolar Cellular Replicon Potency.
J.Med.Chem., 57, 2014
|
|
1KO7
 
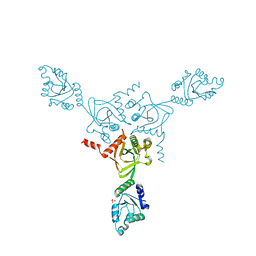 | | X-ray structure of the HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution | | Descriptor: | Hpr kinase/phosphatase, PHOSPHATE ION | | Authors: | Marquez, J.A, Hasenbein, S, Koch, B, Fieulaine, S, Nessler, S, Hengstenberg, W, Scheffzek, K. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the full-length HPr kinase/phosphatase from Staphylococcus xylosus at 1.95 A resolution: Mimicking the product/substrate of the phospho transfer reactions.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KQ8
 
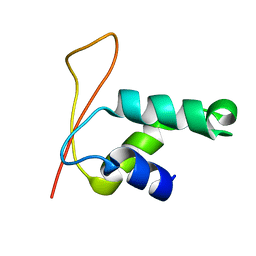 | | Solution Structure of Winged Helix Protein HFH-1 | | Descriptor: | HEPATOCYTE NUCLEAR FACTOR 3 FORKHEAD HOMOLOG 1 | | Authors: | Sheng, W, Rance, M, Liao, X. | | Deposit date: | 2002-01-04 | | Release date: | 2002-01-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure comparison of two conserved HNF-3/fkh proteins HFH-1 and genesis indicates the existence of folding differences in their complexes with a DNA binding sequence.
Biochemistry, 41, 2002
|
|
1DFS
 
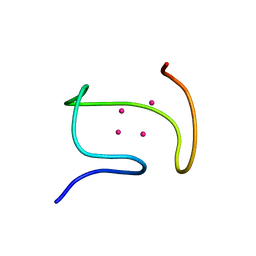 | | SOLUTION STRUCTURE OF THE ALPHA-DOMAIN OF MOUSE METALLOTHIONEIN-1 | | Descriptor: | CADMIUM ION, METALLOTHIONEIN-1 | | Authors: | Zangger, K, Oz, G, Otvos, J.D, Armitage, I.M. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of mouse [Cd7]-metallothionein-1 by homonuclear and heteronuclear NMR spectroscopy.
Protein Sci., 8, 1999
|
|
4RCP
 
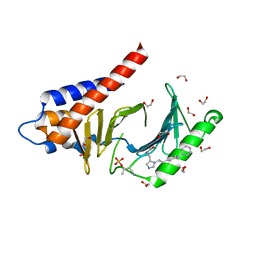 | | Crystal structure of Plk1 Polo-box domain in complex with PL-2 | | Descriptor: | 1,2-ETHANEDIOL, PL-2, Serine/threonine-protein kinase PLK1 | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A new class of peptidomimetics targeting the polo-box domain of polo-like kinase 1.
J.Med.Chem., 58, 2015
|
|
6MBE
 
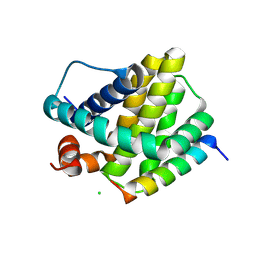 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6MAU
 
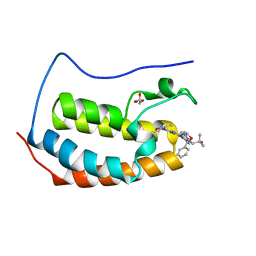 | | Crystal structure of human BRD4(1) in complex with CN210 (compound 19) | | Descriptor: | 1-(4-{6-(3,5-dimethyl-1,2-oxazol-4-yl)-4-[(3S)-3-phenylmorpholin-4-yl]quinazolin-2-yl}-1H-pyrazol-1-yl)-2-methylpropan-2-ol, Bromodomain-containing protein 4, GLYCEROL | | Authors: | Nadupalli, A, Fontano, E, Connors, C.R, Chan, S.G, Olland, A.M, Lakshminarasimhan, D, White, A, Suto, R.K. | | Deposit date: | 2018-08-28 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Lead optimization and efficacy evaluation of quinazoline-based BET family inhibitors for potential treatment of cancer and inflammatory diseases.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
3EUC
 
 | |
4M54
 
 | | The structure of the staphyloferrin B precursor biosynthetic enzyme SbnB bound to N-(1-amino-1-carboxyl-2-ethyl)-glutamic acid and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, N-[(2S)-2-amino-2-carboxyethyl]-L-glutamic acid, ... | | Authors: | Kobylarz, M.J, Murphy, M.E.P. | | Deposit date: | 2013-08-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor.
Chem.Biol., 21, 2014
|
|
1DT4
 
 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
4BZQ
 
 | | Structure of the Mycobacterium tuberculosis APS kinase CysC in complex with ADP and APS | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-PHOSPHOSULFATE, ... | | Authors: | Poyraz, O, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-29 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of the Kinase Domain of the Sulfate-Activating Complex in Mycobacterium Tuberculosis.
Plos One, 10, 2015
|
|
1MV8
 
 | | 1.55 A crystal structure of a ternary complex of GDP-mannose dehydrogenase from Psuedomonas aeruginosa | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, GDP-mannose 6-dehydrogenase, ... | | Authors: | Snook, C.F, Tipton, P.A, Beamer, L.J. | | Deposit date: | 2002-09-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The crystal structure of GDP-mannose
dehydrogenase: A key enzyme in alginate
biosynthesis of P. aeruginosa
Biochemistry, 42, 2003
|
|
4OCZ
 
 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-isobutyrylpiperidin-4-yl)-3-(4-(trifluoromethyl)phenyl)urea | | Descriptor: | 1-[1-(2-methylpropanoyl)piperidin-4-yl]-3-[4-(trifluoromethyl)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morriseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
6CF2
 
 | | Crystal structure of HIV-1 Rev (residues 1-93)-RNA aptamer complex | | Descriptor: | Anti-Rev Antibody, heavy chain, light chain, ... | | Authors: | Eren, E, Dearborn, A.D, Wingfield, P.T. | | Deposit date: | 2018-02-13 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of an RNA Aptamer that Can Inhibit HIV-1 by Blocking Rev-Cognate RNA (RRE) Binding and Rev-Rev Association.
Structure, 26, 2018
|
|
4OLU
 
 | | Crystal structure of antibody VRC07 in complex with clade A/E 93TH057 HIV-1 gp120 core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen binding fragment of heavy chain: Antibody VRC01, Antigen binding fragment of light chain: Antibody VRC01, ... | | Authors: | Kwon, Y.D, Kwong, P.D. | | Deposit date: | 2014-01-25 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo.
J.Virol., 88, 2014
|
|
3EPW
 
 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase in complex with the inhibitor (2R,3R,4S)-1-[(4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-2-(hydroxymethyl)pyrrolidin-3,4-diol | | Descriptor: | 7-(((2R,3R,4S)-3,4-dihydroxy-2-(hydroxymethyl)pyrrolidin-1-yl)methyl)-3H-pyrrolo[3,2-d]pyrimidin-4(5H)-one, CALCIUM ION, IAG-nucleoside hydrolase, ... | | Authors: | Versees, W, Goeminne, A, Berg, M, Vandemeulebroucke, A, Haemers, A, Augustyns, K, Steyaert, J. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors.
Biochim.Biophys.Acta, 1794, 2009
|
|
1M0M
 
 | | BACTERIORHODOPSIN M1 INTERMEDIATE AT 1.43 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, BACTERIORHODOPSIN, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: the switch in the bacteriorhodopsin photocycle.
J.Mol.Biol., 321, 2002
|
|
1M0K
 
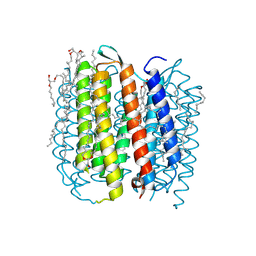 | | BACTERIORHODOPSIN K INTERMEDIATE AT 1.43 A RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, 2,10,23-TRIMETHYL-TETRACOSANE, RETINAL, ... | | Authors: | Lanyi, J.K. | | Deposit date: | 2002-06-13 | | Release date: | 2002-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal.
J.Mol.Biol., 321, 2002
|
|
4OQ9
 
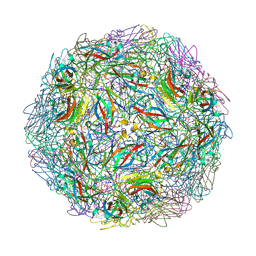 | | Satellite Tobacco Mosaic Virus Refined to 1.4 A Resolution using non-crystallographic symmetry restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
