4MCH
 
 | | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520. | | Descriptor: | 6-hydroxynaphthalene-1-carboxylic acid, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of uridine phosphorylase from vibrio fischeri es114 complexed with 6-hydroxy-1-naphthoic acid, NYSGRC Target 029520.
To be Published
|
|
7DUX
 
 | |
4LSQ
 
 | | Crystal structure of broadly and potently neutralizing antibody VRC-CH31 in complex with HIV-1 clade A/E gp120 93TH057 with LOOP D and Loop V5 from clade A strain 3415_v1_c1 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Multidonor Analysis Reveals Structural Elements, Genetic Determinants, and Maturation Pathway for HIV-1 Neutralization by VRC01-Class Antibodies.
Immunity, 39, 2013
|
|
7WVM
 
 | | The complex structure of PD-1 and cemiplimab | | Descriptor: | Heavy Chain of Cemiplimab, Light Chain of Cemiplimab, Programmed cell death protein 1 | | Authors: | Lu, D, Xu, Z.P, Liu, K.F, Tan, S.G, Gao, G.F, Chai, Y. | | Deposit date: | 2022-02-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | PD-1 N58-Glycosylation-Dependent Binding of Monoclonal Antibody Cemiplimab for Immune Checkpoint Therapy.
Front Immunol, 13, 2022
|
|
4BE4
 
 | | Closed conformation of O. piceae sterol esterase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gutierrez-Fernandez, J, Vaquero, M.E, Prieto, A, Barriuso, J, Gonzalez, M.J, Hermoso, J.A. | | Deposit date: | 2013-03-05 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Ophiostoma Piceae Sterol Esterase: Structural Insights Into Activation Mechanism and Product Release.
J.Struct.Biol., 187, 2014
|
|
6F83
 
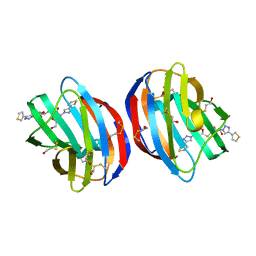 | | Crystal Structure of Human Galectin-1 in Complex With Thienyl-1,2, 3-triazolyl Thiodigalactoside Inhibitor | | Descriptor: | 3-deoxy-3-[4-(thiophen-3-yl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-1-thio-3-[4-(thiophen-3-yl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranoside, Galectin-1 | | Authors: | Collins, P.M, Blanchard, H. | | Deposit date: | 2017-12-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aromatic heterocycle galectin-1 interactions for selective single-digit nM affinity ligands
To be published
|
|
4Z1Q
 
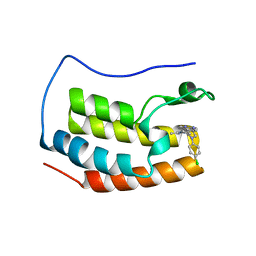 | | Crystal structure of the first bromodomain of human BRD4 bound to benzotriazolo-diazepine scaffold | | Descriptor: | 5-[(4R)-6-(4-chlorophenyl)-1,4-dimethyl-5,6-dihydro-4H-[1,2,4]triazolo[4,3-a][1,5]benzodiazepin-8-yl]pyridin-2-amine, Bromodomain-containing protein 4 | | Authors: | Setser, J.W, Poy, F, Tang, Y, Bellon, S.F. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Discovery of Benzotriazolo[4,3-d][1,4]diazepines as Orally Active Inhibitors of BET Bromodomains.
Acs Med.Chem.Lett., 7, 2016
|
|
6MGU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate
To Be Published
|
|
4J3J
 
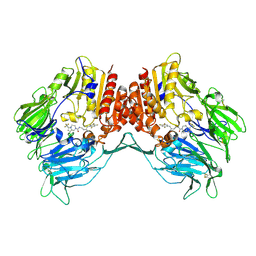 | | Crystal Structure of DPP-IV with Compound C3 | | Descriptor: | Dipeptidyl peptidase 4, N-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butyl]-6-(trifluoromethyl)-3,4-dihydropyrrolo[1,2-a]pyrazine-2(1H)-carboxamide | | Authors: | Xiong, B, Zhu, L.R, Chen, D.Q, Zhao, Y.L, Jiang, F, Shen, J.K. | | Deposit date: | 2013-02-05 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design and synthesis of 4-(2,4,5-trifluorophenyl)butane-1,3-diamines as dipeptidyl peptidase IV inhibitors
Chemmedchem, 8, 2013
|
|
4P7N
 
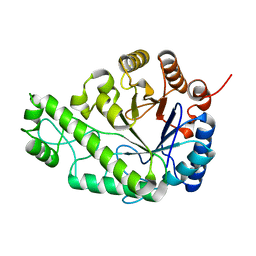 | | Structure of Escherichia coli PgaB C-terminal domain in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase | | Authors: | Little, D.J, Li, G, Ing, C, DiFrancesco, B, Bamford, N.C, Robinson, H, Nitz, M, Pomes, R, Howell, P.L. | | Deposit date: | 2014-03-27 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Modification and periplasmic translocation of the biofilm exopolysaccharide poly-beta-1,6-N-acetyl-D-glucosamine.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1QDT
 
 | |
1FGY
 
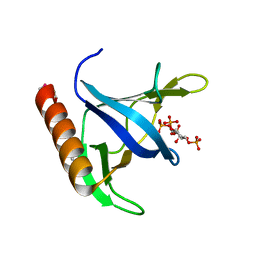 | | GRP1 PH DOMAIN WITH INS(1,3,4,5)P4 | | Descriptor: | GRP1, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Lietzke, S.E, Bose, S, Cronin, T, Klarlund, J, Chawla, A, Czech, M.P, Lambright, D.G. | | Deposit date: | 2000-07-29 | | Release date: | 2000-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains.
Mol.Cell, 6, 2000
|
|
5ZJ8
 
 | |
1H0C
 
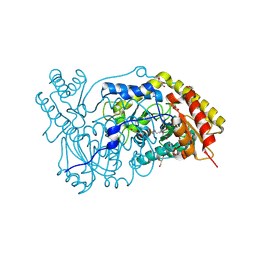 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
6M9L
 
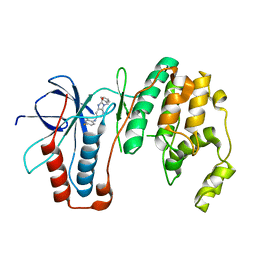 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridine-2-one based p38 MAP Kinase Inhibitors by scaffold hopping - compound 10 | | Descriptor: | 3-benzyl-6-[(2,4-difluorophenyl)amino]-1,3-dihydro-2H-imidazo[4,5-b]pyridin-2-one, Mitogen-activated protein kinase 14 | | Authors: | Lane, W, Okada, K. | | Deposit date: | 2018-08-23 | | Release date: | 2019-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Based Design, Synthesis, and Biological Evaluation of Imidazo[4,5-b]pyridin-2-one-Based p38 MAP Kinase Inhibitors: Part 1.
Chemmedchem, 14, 2019
|
|
6BMX
 
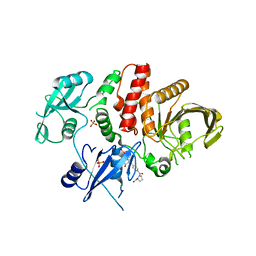 | | Non-receptor Protein Tyrosine Phosphatase SHP2 in Complex with Allosteric Inhibitor SHP844 | | Descriptor: | 1-(3-chloro-4-{[1-(2-hydroxy-3-methoxyphenyl)-5-oxo[1,2,4]triazolo[4,3-a]quinazolin-4(5H)-yl]methyl}benzene-1-carbonyl)-L-proline, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stams, T, Fodor, M. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Dual Allosteric Inhibition of SHP2 Phosphatase.
ACS Chem. Biol., 13, 2018
|
|
6LRB
 
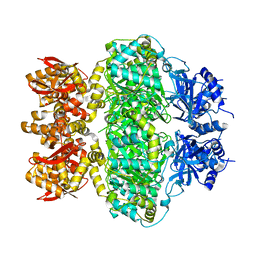 | |
4YDV
 
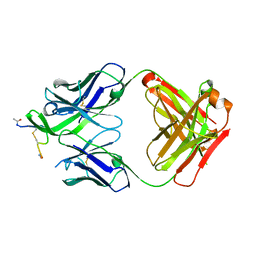 | | STRUCTURE OF THE ANTIBODY 7B2 THAT CAPTURES HIV-1 VIRIONS | | Descriptor: | HIV ANTIBODY 7B2 HEAVY CHAIN,IgG H chain, HIV ANTIBODY 7B2 LIGHT CHAIN,Ig kappa chain C region, HIV GP41 PEPTIDE GP41(596-606) | | Authors: | Nicely, N.I, Pemble IV, C.W. | | Deposit date: | 2015-02-23 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human Non-neutralizing HIV-1 Envelope Monoclonal Antibodies Limit the Number of Founder Viruses during SHIV Mucosal Infection in Rhesus Macaques.
Plos Pathog., 11, 2015
|
|
3OHG
 
 | |
4NKA
 
 | | Crystal structure of human fibroblast growth factor receptor 1 kinase domain in complex with pyrazolaminopyrimidine 2 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N~4~-{3-[2-(3,4-dimethoxyphenyl)ethyl]-1H-pyrazol-5-yl}-N~2~-[(3-methyl-1,2-oxazol-5-yl)methyl]pyrimidine-2,4-diamine, ... | | Authors: | Norman, R.A, Klein, T. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | FGFR1 Kinase Inhibitors: Close Regioisomers Adopt Divergent Binding Modes and Display Distinct Biophysical Signatures.
ACS Med Chem Lett, 5, 2014
|
|
1XM9
 
 | |
3MK8
 
 | |
7XIW
 
 | | SARS-CoV-2 Omicron BA.2 variant spike (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-04-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection.
Nature, 608, 2022
|
|
4NIA
 
 | | Satellite Tobacco Mosaic Virus Refined at room temperature to 1.8 A Resolution using NCS Restraints | | Descriptor: | Coat protein, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Larson, S.B, Day, J.S, McPherson, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Satellite tobacco mosaic virus refined to 1.4 angstrom resolution.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5V80
 
 | |
