7N73
 
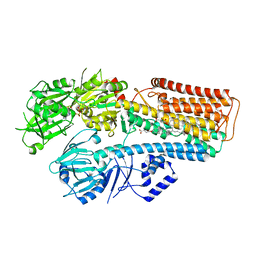 | |
7N72
 
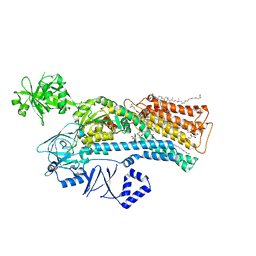 | |
7N74
 
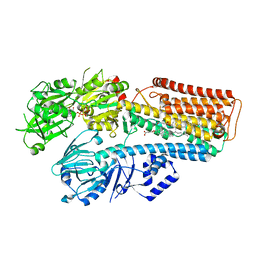 | |
7N70
 
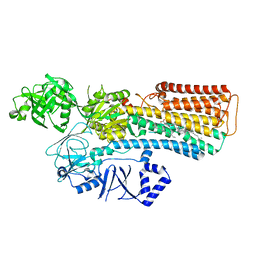 | |
7N78
 
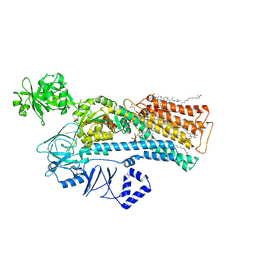 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sim, S.I, Park, E. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-10 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of polyamine transport by human ATP13A2 (PARK9).
Mol.Cell, 81, 2021
|
|
4XQW
 
 | | X-ray structure analysis of xylanase-N44E with MES at pH6.0 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-20 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7N77
 
 | |
6PLZ
 
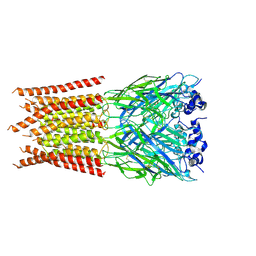 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, closed state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
5MPE
 
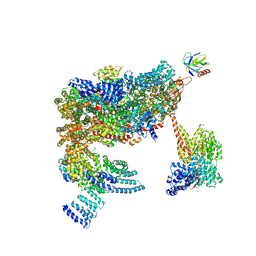 | | 26S proteasome in presence of ATP (s2) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MEC
 
 | |
6PLY
 
 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with GABA in SMA, open state | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, Glycine receptor subunit alphaZ1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
4XPV
 
 | | Neutron and X-ray structure analysis of xylanase: N44D at pH6 | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-18 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6P4P
 
 | | Salmonella typhi PltB Homopentamer N29K Mutant | | Descriptor: | Putative pertussis-like toxin subunit | | Authors: | Nguyen, T, Milano, S.K, Hillpot, E.C, Yang, Y.A, Song, J. | | Deposit date: | 2019-05-28 | | Release date: | 2020-03-25 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Cell Host Microbe, 27, 2020
|
|
7JYE
 
 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | Descriptor: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
7NHF
 
 | | Crystal structure of Arabidopsis thaliana Pdx1K166R | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
6PRZ
 
 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
8IQ4
 
 | | Cryo-EM structure of Carboprost-bound prostaglandin-F2-alpha receptor-miniGq-Nb35 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Lv, X, Gao, K, Nie, J, Zhang, X, Zhang, S, Ren, Y, Li, Q, Huang, J, Liu, L, Zhang, X, Sun, X, Zhang, W, Liu, X. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of human prostaglandin F 2 alpha receptor reveal the mechanism of ligand and G protein selectivity.
Nat Commun, 14, 2023
|
|
6P8P
 
 | | Structure of P. aeruginosa ATCC27853 HORMA1 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
5MDI
 
 | | Crystal structure of TDP-43 N-terminal domain at 2.1 A resolution | | Descriptor: | ACETATE ION, TAR DNA-binding protein 43 | | Authors: | Afroz, T, Hock, E.-M, Ernst, P, Foglieni, C, Jambeau, M, Gilhespy, L, Laferriere, F, Maniecka, Z, Plueckthun, A, Mittl, P, Paganetti, P, Allain, F.H.T, Polymenidou, M. | | Deposit date: | 2016-11-11 | | Release date: | 2017-07-05 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and dynamic polymerization of the ALS-linked protein TDP-43 antagonizes its pathologic aggregation.
Nat Commun, 8, 2017
|
|
5EUY
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
1IRS
 
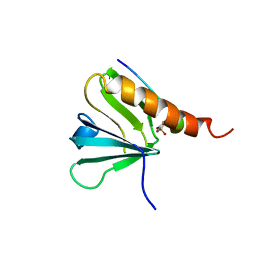 | | IRS-1 PTB DOMAIN COMPLEXED WITH A IL-4 RECEPTOR PHOSPHOPEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | IL-4 RECEPTOR PHOSPHOPEPTIDE, IRS-1 | | Authors: | Zhou, M.-M, Huang, B, Olejniczak, E.T, Meadows, R.P, Shuker, S.B, Miyazaki, M, Trub, T, Shoelson, S.E, Feisk, S.W. | | Deposit date: | 1996-03-22 | | Release date: | 1997-05-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural basis for IL-4 receptor phosphopeptide recognition by the IRS-1 PTB domain.
Nat.Struct.Biol., 3, 1996
|
|
7N8L
 
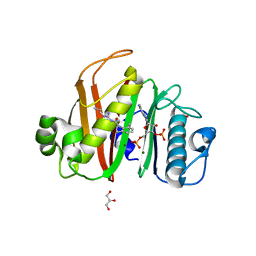 | |
5MRD
 
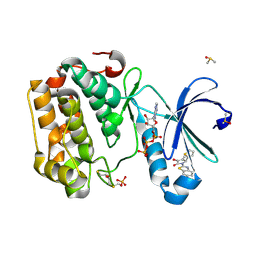 | | Human PDK1-PKCiota Kinase Chimera in Complex with Allosteric Compound PS267 Bound to the PIF-Pocket | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-phosphoinositide-dependent protein kinase 1, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Arencibia, J.M, Froehner, W, Krupa, M, Pastor-Flores, D, Merker, P, Oellerich, T, Neimanis, S, Schmithals, C, Koeberle, V, Suess, E, Zeuzem, S, Stark, H, Piiper, A, Odadzic, D, Schulze, J.O, Biondi, R.M. | | Deposit date: | 2016-12-22 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | An Allosteric Inhibitor Scaffold Targeting the PIF-Pocket of Atypical Protein Kinase C Isoforms.
ACS Chem. Biol., 12, 2017
|
|
4E1V
 
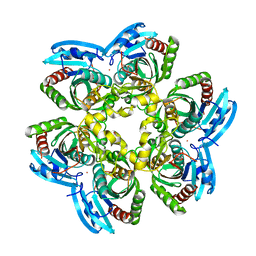 | | X-RAY Structure of the Uridine Phosphorylase from Salmonella Typhimurium in Complex with 5-Fluorouracil at 2.15 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, 5-FLUOROURACIL, GLYCEROL, ... | | Authors: | Lashkov, A.A, Sotnichenko, S.E, Prokofev, I.I, Gabdoulkhakov, A.G, Mikhailov, A.M. | | Deposit date: | 2012-03-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray structure of Salmonella typhimurium uridine phosphorylase complexed with 5-fluorouracil and molecular modelling of the complex of 5-fluorouracil with uridine phosphorylase from Vibrio cholerae.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
7MZV
 
 | | Structure of yeast pseudouridine synthase 7 (PUS7) | | Descriptor: | Multisubstrate pseudouridine synthase 7, SULFATE ION | | Authors: | Purchal, M, Koutmos, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Pseudouridine synthase 7 is an opportunistic enzyme that binds and modifies substrates with diverse sequences and structures.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
