6XQV
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | Descriptor: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQZ
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
 | |
6XQX
 
 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | Authors: | Fenton, B.A, Zhou, P, Davies, C. | | Deposit date: | 2020-07-10 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
5BOH
 
 | |
6XJ3
 
 | | Crystal structure of Class D beta-lactamase from Klebsiella quasipneumoniae in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Chang, C, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Class D beta-lactamase from Klebsiella quasipneumoniae
To Be Published
|
|
6SKP
 
 | |
6SKR
 
 | | OXA-10_ETP. Structural insight to the enhanced carbapenem efficiency of OXA-655 compared to OXA-10. | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, FORMIC ACID, ... | | Authors: | Leiros, H.-K.S. | | Deposit date: | 2019-08-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10.
Febs Open Bio, 10, 2020
|
|
6SKQ
 
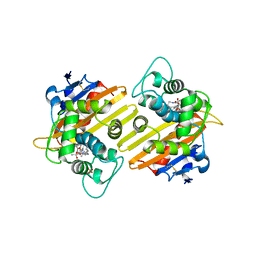 | |
6T1H
 
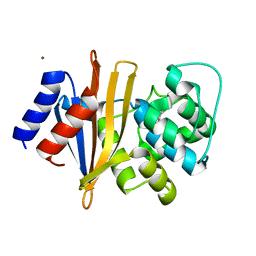 | | OXA-51-like beta-lactamase OXA-66 | | Descriptor: | Beta-lactamase OXA-66, ZINC ION | | Authors: | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
3ZNT
 
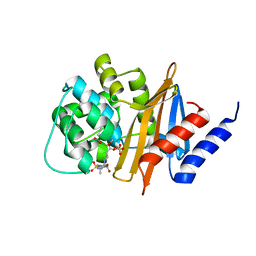 | | Crystal structure of OXA-24 class D beta-lactamase with tazobactam | | Descriptor: | BETA-LACTAMASE, SULFATE ION, TAZOBACTAM INTERMEDIATE | | Authors: | Power, P, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2013-02-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Oxa-24 Beta-Lactamase Inhibited by Tazobactam
To be Published
|
|
6RTN
 
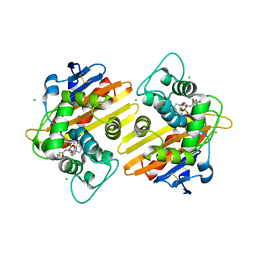 | | Crystal structure of OXA-10 with VNRX-5133 | | Descriptor: | (3~{R})-3-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Beta-lactamase OXA-10, CHLORIDE ION | | Authors: | Brem, J, Schofield, C. | | Deposit date: | 2019-05-24 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-beta-Lactamases.
J.Med.Chem., 62, 2019
|
|
6WHL
 
 | |
6XQR
 
 | | OXA-48 bound by Compound 2.2 | | Descriptor: | Beta-lactamase, CHLORIDE ION, [1,1'-biphenyl]-4,4'-disulfonic acid | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-07-10 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
6WY4
 
 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-12 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
5CTN
 
 | | Structure of BPu1 beta-lactamase | | Descriptor: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, CITRATE ANION | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-25 | | Last modified: | 2016-10-05 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5CRF
 
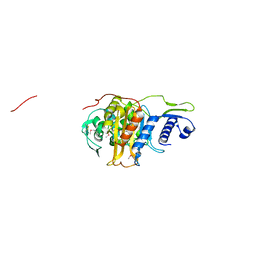 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5CTM
 
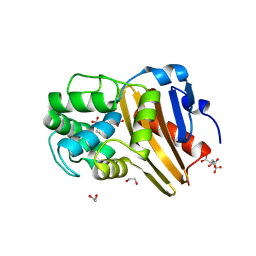 | | Structure of BPu1 beta-lactamase | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CITRATE ANION, ... | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-07-24 | | Release date: | 2015-11-18 | | Last modified: | 2015-12-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Class D beta-lactamases do exist in Gram-positive bacteria.
Nat.Chem.Biol., 12, 2016
|
|
5KZH
 
 | |
8SQF
 
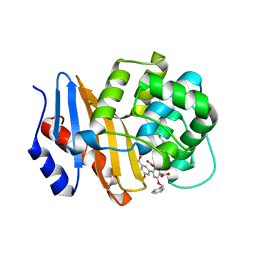 | | OXA-48 bound to inhibitor CDD-2725 | | Descriptor: | (1M)-3'-(benzyloxy)-5-hydroxy[1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
8SQG
 
 | | OXA-48 bound to inhibitor CDD-2801 | | Descriptor: | (1M)-3'-(benzyloxy)-5-[2-(methylamino)-2-oxoethoxy][1,1'-biphenyl]-3,4'-dicarboxylic acid, BICARBONATE ION, Beta-lactamase | | Authors: | Park, S, Judge, A, Fan, J, Sankaran, B, Palzkill, T. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploiting the Carboxylate-Binding Pocket of beta-Lactamase Enzymes Using a Focused DNA-Encoded Chemical Library.
J.Med.Chem., 67, 2024
|
|
5DTK
 
 | | Fragments bound to the OXA-48 beta-lactamase: Compound 17 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-di(pyridin-4-yl)benzoic acid, Beta-lactamase, ... | | Authors: | Lund, B.A, Christopeit, T, Leiros, H.-K.S. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.60000241 Å) | | Cite: | Screening and Design of Inhibitor Scaffolds for the Antibiotic Resistance Oxacillinase-48 (OXA-48) through Surface Plasmon Resonance Screening.
J.Med.Chem., 59, 2016
|
|
5L2F
 
 | | High Resolution Structure of Acinetobacter baumannii beta-lactamase OXA-51 I129L/K83D bound to doripenem | | Descriptor: | (4R,5S)-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-3-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-4,5-dihydro-1H-pyrrole-2-carboxylic acid, ACETATE ION, Beta-lactamase, ... | | Authors: | June, C.M, Powers, R.A, Leonard, D.A, Muckenthaler, T. | | Deposit date: | 2016-08-01 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of a doripenem-bound OXA-51 class D beta-lactamase variant with enhanced carbapenemase activity.
Protein Sci., 25, 2016
|
|
5FQ9
 
 | | Crystal structure of the OXA10 with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, ACETATE ION, BETA-LACTAMASE OXA-10, ... | | Authors: | Brem, J, McDonough, M.A, Cain, R, Clifton, I, Fishwick, C.W.G, Schofield, C.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of metallo-beta-lactamase, serine-beta-lactamase and penicillin-binding protein inhibition by cyclic boronates.
Nat Commun, 7, 2016
|
|
5MMY
 
 | | Crystal structure of OXA10 with HEPES | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase OXA-10, ... | | Authors: | Brem, J. | | Deposit date: | 2016-12-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | (13)C-Carbamylation as a mechanistic probe for the inhibition of class D beta-lactamases by avibactam and halide ions.
Org. Biomol. Chem., 15, 2017
|
|
