6BZB
 
 | |
4GLS
 
 | | Crystal Structure of Chemically Synthesized Heterochiral {D-Protein Antagonist plus VEGF-A} Protein Complex in space group P21 | | Descriptor: | D- RFX001, D- Vascular endothelial growth factor-A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mandal, K, Uppalapati, M, Ault-Riche, D, Kenney, J, Lowitz, J, Sidhu, S, Kent, S.B.H. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical synthesis and X-ray structure of a heterochiral {D-protein antagonist plus vascular endothelial growth factor} protein complex by racemic crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6ZBR
 
 | | VEGF-A 13:107 crystallized with 4C bicyclic peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Chains: P, PHOSPHATE ION, ... | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
6ZFL
 
 | | High resolution structure of VEGF-A 12:107 crystallized in tetragonal form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHATE ION, Vascular endothelial growth factor A | | Authors: | Gaucher, J.-F, Broussy, S, Reille-Seroussi, M. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and ITC Characterization of Peptide-Protein Binding: Thermodynamic Consequences of Cyclization Constraints, a Case Study on Vascular Endothelial Growth Factor Ligands.
Chemistry, 2022
|
|
2GJ4
 
 | | Structure of rabbit muscle glycogen phosphorylase in complex with ligand | | Descriptor: | (5-HYDROXY-4,6-DIMETHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, 2-CHLORO-N-[(1R,2R)-1-HYDROXY-2,3-DIHYDRO-1H-INDEN-2-YL]-6H-THIENO[2,3-B]PYRROLE-5-CARBOXAMIDE, Glycogen phosphorylase, ... | | Authors: | Otterbein, L.R, Pannifer, A.D, Tucker, J, Breed, J, Oikonomakos, N.G, Rowsell, S, Pauptit, R.A, Claire, M. | | Deposit date: | 2006-03-30 | | Release date: | 2007-02-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel thienopyrrole glycogen phosphorylase inhibitors: synthesis, in vitro SAR and crystallographic studies.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1U0F
 
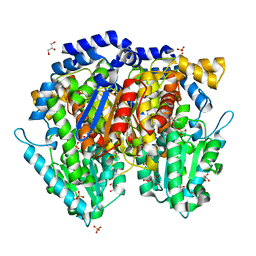 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
1U0E
 
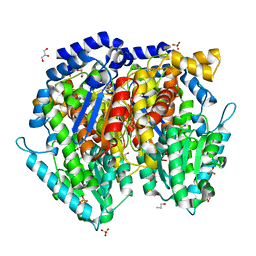 | | Crystal structure of mouse phosphoglucose isomerase | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, Glucose-6-phosphate isomerase, ... | | Authors: | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2004-07-13 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
4X39
 
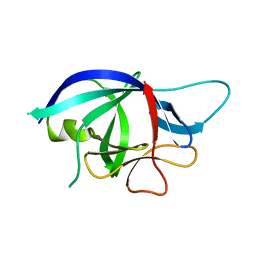 | |
5QQ9
 
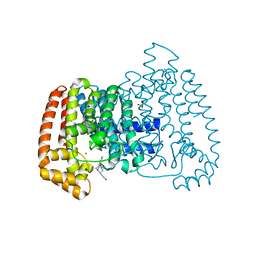 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000567a | | Descriptor: | 2-[(2R,5R,6S)-5-hydroxy-1,2,4,5,6,7-hexahydro-3H-2,6-methanoazocino[5,4-b]indol-3-yl]acetamide, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QPS
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000644a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ7
 
 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000562a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
3O4W
 
 | | Crystal structure of dimeric KlHxk1 in crystal form IV | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Hexokinase, ... | | Authors: | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | Deposit date: | 2010-07-27 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
6R0B
 
 | | T. cruzi FPPS in complex with 3-((4-(5-chlorobenzo[d]thiazol-2-yl)piperazin-1-yl)methyl)-1H-indol-5-ol | | Descriptor: | 3-[[4-(5-chloranyl-1,3-benzothiazol-2-yl)piperazin-1-yl]methyl]-1~{H}-indol-5-ol, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Muenzker, L, Schleberger, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Targeting farnesyl pyrophosphate synthase of Trypanosoma cruzi by fragment-based lead discovery
Thesis, 2019
|
|
5QQC
 
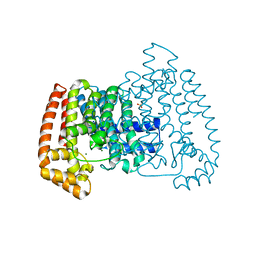 | | Crystal Structure of T. cruzi FPPS after initial refinement with no ligand modelled (structure $n) | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, MAGNESIUM ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition of ground-state model - FPPS screened against the DSI Fragment Library
To Be Published
|
|
6SF8
 
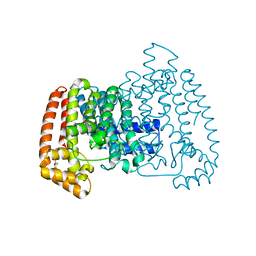 | | Trypanosoma cruzi farnesyl diphosphate synthase in complex with fragment j51 | | Descriptor: | 3-[(4E)-4-imino-5,6-dimethylfuro[2,3-d]pyrimidin-3(4H)-yl]-N,N-dimethylpropan-1-amine, Farnesyl diphosphate synthase, SULFATE ION | | Authors: | Magari, F, Heine, A, Klebe, G. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Trypanosoma cruzi farnesyl diphosphate synthase in complex with fragment j51
To Be Published
|
|
5QPZ
 
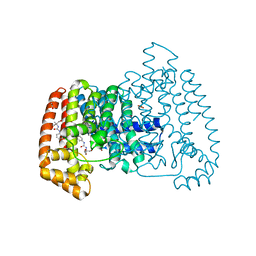 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000524a | | Descriptor: | 1-[2-methyl-1,3-bis(oxidanyl)propan-2-yl]-3-phenyl-urea, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ8
 
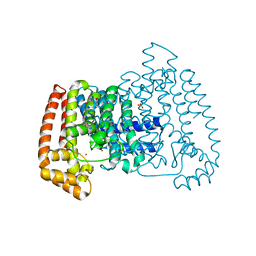 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000563a | | Descriptor: | 3-ethyl-1-[(1~{R},8~{S},9~{S},10~{S})-10-oxidanyl-11-oxatricyclo[6.2.1.0^{2,7}]undeca-2(7),3,5-trien-9-yl]imidazolidine-2,4-dione, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
1IAT
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHOGLUCOSE ISOMERASE/NEUROLEUKIN/AUTOCRINE MOTILITY FACTOR/MATURATION FACTOR | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHOGLUCOSE ISOMERASE, SULFATE ION | | Authors: | Read, J.A, Pearce, J, Li, X, Muirhead, H, Chirgwin, J, Davies, C. | | Deposit date: | 2001-03-23 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The crystal structure of human phosphoglucose isomerase at 1.6 A resolution: implications for catalytic mechanism, cytokine activity and haemolytic anaemia.
J.Mol.Biol., 309, 2001
|
|
5R8J
 
 | |
4X37
 
 | |
5GHU
 
 | | Crystal structure of YadV chaperone at 1.63 Angstrom | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Pandey, N.K, Bhavesh, N.S. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of the usher chaperone YadV reveals a monomer with the proline lock in closed conformation suggestive of an intermediate state.
Febs Lett., 2020
|
|
8CGK
 
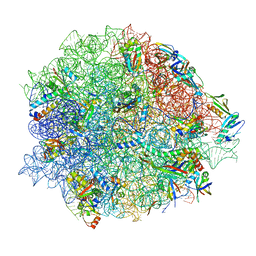 | | Lincomycin and Avilamycin bound to the 50S subunit | | Descriptor: | (2R,3S,4R,6S)-4-hydroxy-6-{[(2R,3aR,4R,4'R,5'S,6S,6'R,7aR)-4'-hydroxy-6-{[(2S,3R,4R,5S,6R)-3-hydroxy-2-{[(2R,3S,4S,5S,6S)-4-hydroxy-6-({(2R,3aS,3a'R,6S,6'R,7R,7'R,7aR,7a'R)-7'-hydroxy-7'-[(1S)-1-hydroxyethyl]-6'-methyl-7-[(2-methylpropanoyl)oxy]octahydro-4H-2,4'-spirobi[[1,3]dioxolo[4,5-c]pyran]-6-yl}oxy)-5-methoxy-2-(methoxymethyl)tetrahydro-2H-pyran-3-yl]oxy}-5-methoxy-6-methyltetrahydro-2H-pyran-4-yl]oxy}-4,6',7a-trimethyloctahydro-4H-spiro[1,3-dioxolo[4,5-c]pyran-2,2'-pyran]-5'-yl]oxy}-2-methyltetrahydro-2H-pyran-3-yl 3,5-dichloro-4-hydroxy-2-methoxy-6-methylbenzoate (non-preferred name), 23S rRNA, 50S ribosomal protein L15, ... | | Authors: | Paternoga, H, Crowe-McAuliffe, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2023-02-05 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.64 Å) | | Cite: | Structural conservation of antibiotic interaction with ribosomes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5QTD
 
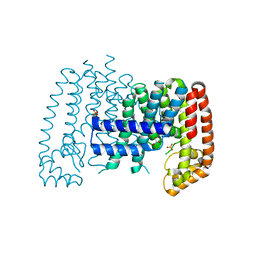 | | Unliganded T. brucei FPPS | | Descriptor: | Farnesyl pyrophosphate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Muenzker, L, Petrick, J.K, Schleberger, C, Cornaciu, I, Marquez, J.A, Jahnke, W. | | Deposit date: | 2019-08-09 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Unliganded T. brucei FPPS
To Be Published
|
|
5QTC
 
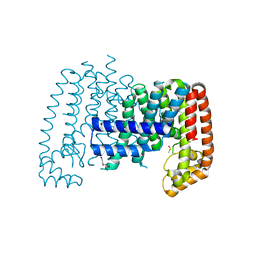 | |
2CXU
 
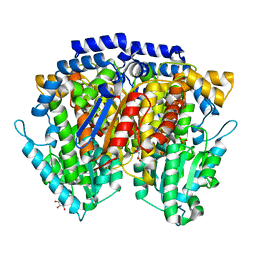 | | Crystal structure of mouse AMF / M6P complex | | Descriptor: | GLYCEROL, Glucose-6-phosphate isomerase, PHOSPHATE ION | | Authors: | Tanaka, N, Haga, A, Naba, N, Shiraiwa, K, Kusakabe, Y, Hashimoto, K, Funasaka, T, Nagase, H, Raz, A, Nakamura, K.T. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of mouse autocrine motility factor in complex with carbohydrate phosphate inhibitors provide insight into structure-activity relationship of the inhibitors
J.Mol.Biol., 356, 2006
|
|
