6OXT
 
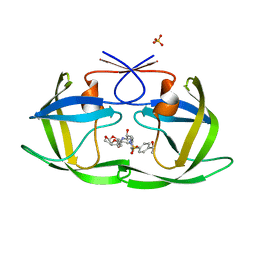 | | HIV-1 Protease NL4-3 WT in Complex with LR-84 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OY0
 
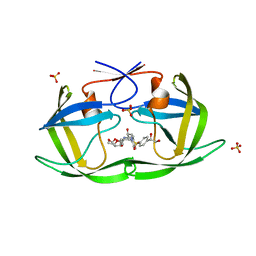 | | HIV-1 Protease NL4-3 WT in Complex with LR2-21 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-4-{({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)[(2S)-2-methylbutyl]amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OTG
 
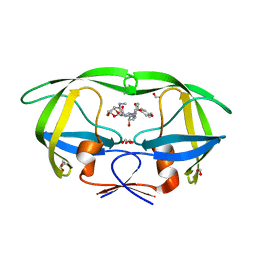 | | HIV-1 protease triple mutants V32I, I47V, V82I with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, FORMIC ACID, GLYCEROL, ... | | Authors: | Wang, Y.-F, Pawar, S, Weber, I.T. | | Deposit date: | 2019-05-03 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural studies of antiviral inhibitor with HIV-1 protease bearing drug resistant substitutions of V32I, I47V and V82I.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
6OXW
 
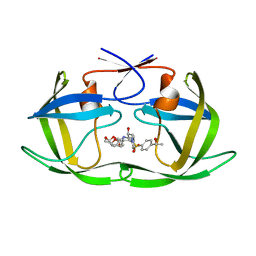 | | HIV-1 Protease NL4-3 WT in Complex with LR-100 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1R)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
5VDN
 
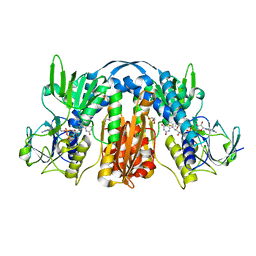 | | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Glutathione oxidoreductase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-03 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Glutathione Reductase from Yersinia pestis in Complex with FAD.
To Be Published
|
|
6OXQ
 
 | | HIV-1 Protease NL4-3 WT in Complex with UMass8 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-3-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-hydroxypropyl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXV
 
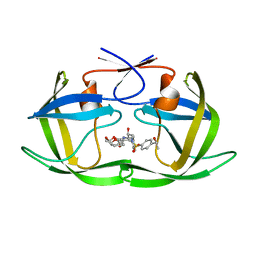 | | HIV-1 Protease NL4-3 WT in Complex with LR-85 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[(2-ethylbutyl)({4-[(1S)-1-hydroxyethyl]phenyl}sulfonyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXX
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-18 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1R)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3 | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
6OXY
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR2-19 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-[({4-[(1S)-1,2-dihydroxyethyl]phenyl}sulfonyl)(2-methylpropyl)amino]-3-hydroxy-1-phenylbutan-2-yl}carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Lee, S.K, Henes, M, Kosovrasti, K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | HIV-1 Protease Inhibitors Incorporating Stereochemically Defined P2' Ligands To Optimize Hydrogen Bonding in the Substrate Envelope.
J.Med.Chem., 62, 2019
|
|
5VEW
 
 | | Structure of the human GLP-1 receptor complex with PF-06372222 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Glucagon-like peptide 1 receptor,Endolysin chimera, N-{4-[(R)-(3,3-dimethylcyclobutyl)({6-[4-(trifluoromethyl)-1H-imidazol-1-yl]pyridin-3-yl}amino)methyl]benzene-1-carbonyl}-beta-alanine, ... | | Authors: | Song, G, Yang, D, Wang, Y, Graaf, C.D, Zhou, Q, Jiang, S, Liu, K, Cai, X, Dai, A, Lin, G, Liu, D, Wu, F, Wu, Y, Zhao, S, Ye, L, Han, G.W, Lau, J, Wu, B, Hanson, M.A, Liu, Z.-J, Wang, M.-W, Stevens, R.C. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators.
Nature, 546, 2017
|
|
5VP0
 
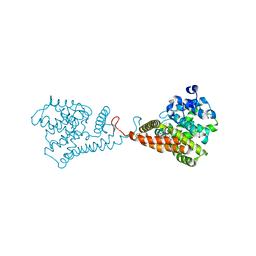 | | Discovery of Clinical Candidate N-{(1S)-1-[3-Fluoro-4-(trifluoromethoxy)phenyl]-2-methoxyethyl}-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915), A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders | | Descriptor: | MAGNESIUM ION, N-{(1S)-1-[3-fluoro-4-(trifluoromethoxy)phenyl]-2-methoxyethyl}-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, ZINC ION, ... | | Authors: | Hoffman, I.D. | | Deposit date: | 2017-05-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Clinical Candidate N-((1S)-1-(3-Fluoro-4-(trifluoromethoxy)phenyl)-2-methoxyethyl)-7-methoxy-2-oxo-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide (TAK-915): A Highly Potent, Selective, and Brain-Penetrating Phosphodiesterase 2A Inhibitor for the Treatment of Cognitive Disorders.
J. Med. Chem., 60, 2017
|
|
5VPG
 
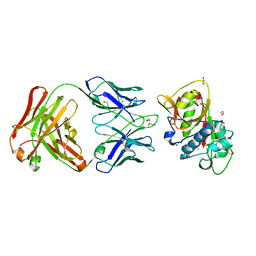 | | CRYSTAL STRUCTURE OF DER P 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
5VPL
 
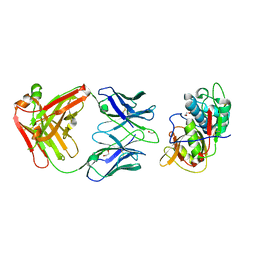 | | CRYSTAL STRUCTURE OF DER F 1 COMPLEXED WITH FAB 4C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4C1 - HEAVY CHAIN, ... | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Determinants For Antibody Binding On Group 1 House Dust Mite Allergens.
J.Biol.Chem., 287, 2012
|
|
7N3Y
 
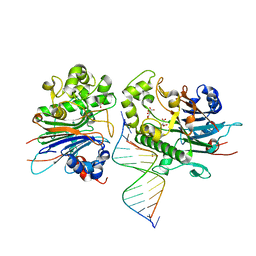 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain E59Q/D222N Mutant in Complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CITRIC ACID, ... | | Authors: | Wojtaszek, J.L, Krahn, J, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
5VRM
 
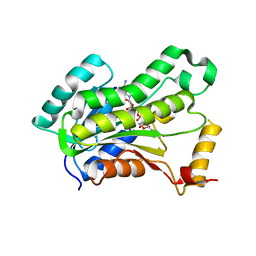 | | CRYSTAL STRUCTURE OF THE INHA FROM MYCOBACTERIUM TUBERCULOSIS IN COMPLEX WITH AN12855, EBSI 4333. | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], [[(2~{R},3~{S},4~{R},5~{R})-5-(3-aminocarbonylpyridin-1-ium-1-yl)-3-oxidanyl-4-[[1-oxidanyl-6-[4-(trifluoromethyl)phenoxy]-3~{H}-2,1$l^{4}-benzoxaborol-1-yl]oxy]oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Abendroth, J, Edwards, T.E, Lorimer, D. | | Deposit date: | 2017-05-11 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a cofactor-independent inhibitor ofMycobacterium tuberculosisInhA.
Life Sci Alliance, 1, 2018
|
|
7N3Z
 
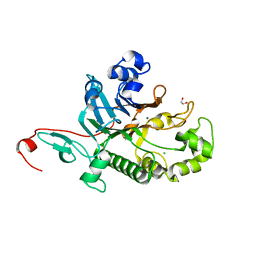 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
3SNO
 
 | |
3CV0
 
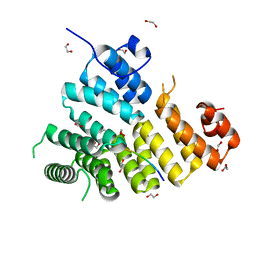 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3CVN
 
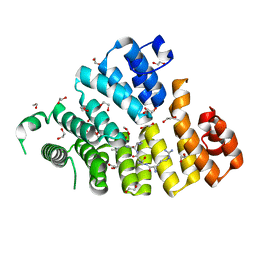 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor, T. brucei GAPDH PTS1 peptide Ac-DRDAAKL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-18 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
3JSM
 
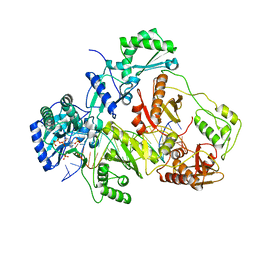 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with tenofovir-diphosphate as the incoming nucleotide substrate | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3SFG
 
 | |
3CYW
 
 | | Effect of Flap Mutations on Structure of HIV-1 Protease and Inhibition by Saquinavir and Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-04-27 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3D1X
 
 | | Crystal structure of HIV-1 mutant I54M and inhibitor saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
