7S7D
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH SYNTHETIC SULFO-MLL PEPTIDE ANALOG | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovska, L, Patskovsky, Y, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-15 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7S8E
 
 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4BCH
 
 | | Structure of CDK9 in complex with cyclin T and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 4-(4-methyl-2-methylimino-3H-1,3-thiazol-5-yl)-2-[(4-methyl-3-morpholin-4-ylsulfonyl-phenyl)amino]pyrimidine-5-carbonitrile, CYCLIN-DEPENDENT KINASE 9, CYCLIN-T1, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-09 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.958 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
6G2S
 
 | | Crystal structure of FimH in complex with a pentaflourinated biphenyl alpha D-mannoside | | Descriptor: | (2~{R},3~{S},4~{S},5~{S},6~{R})-2-(hydroxymethyl)-6-[4-[2,3,4,5,6-pentakis(fluoranyl)phenyl]phenoxy]oxane-3,4,5-triol, SULFATE ION, Type 1 fimbrin D-mannose specific adhesin | | Authors: | Jakob, R.P, Schoenemann, W, Cramer, J, Muehlethaler, T, Daetwyler, P, Zihlmann, P, Fiege, B, Sager, C.P, Smiesko, M, Rabbani, S, Eris, D, Schwardt, O, Maier, T, Ernst, B. | | Deposit date: | 2018-03-23 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improvement of Aglycone pi-Stacking Yields Nanomolar to Sub-nanomolar FimH Antagonists.
Chemmedchem, 14, 2019
|
|
7S8F
 
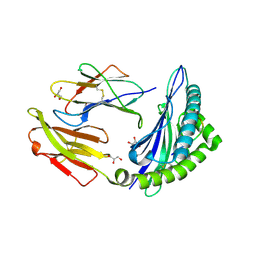 | | STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PEPTIDE AND BOUND GLYCEROL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Patskovsky, Y, Nyovanie, S, Patskovska, L, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-09-17 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
6G4P
 
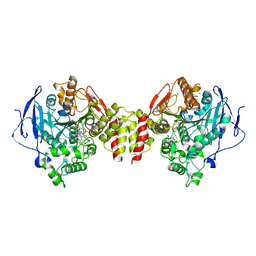 | | Non-aged form of Torpedo californica acetylcholinesterase inhibited by tabun analog NEDPA bound to uncharged reactivator 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[4-[(7-chloranyl-1,2,3,4-tetrahydroacridin-9-yl)amino]butyl]-2-[(~{Z})-hydroxyiminomethyl]pyridin-3-ol, Acetylcholinesterase, ... | | Authors: | Santoni, G, De la Mora, E, de Souza, J, Silman, I, Sussman, J, Baati, R, Weik, M, Nachon, F. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure-Based Optimization of Nonquaternary Reactivators of Acetylcholinesterase Inhibited by Organophosphorus Nerve Agents.
J. Med. Chem., 61, 2018
|
|
4NI1
 
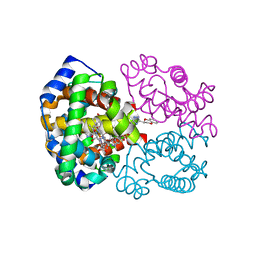 | | Quaternary R CO-liganded hemoglobin structure in complex with a thiol containing compound | | Descriptor: | 5-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-2,4-dihydro-3H-1,2,4-triazole-3-thione, CARBON MONOXIDE, Hemoglobin subunit alpha, ... | | Authors: | Safo, M.K, Meadows, J, Ko, T.-P, Nakagawa, A, Zapol, W. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a Small Molecule that Increases Hemoglobin Oxygen Affinity and Reduces SS Erythrocyte Sickling.
Acs Chem.Biol., 9, 2014
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
4NK3
 
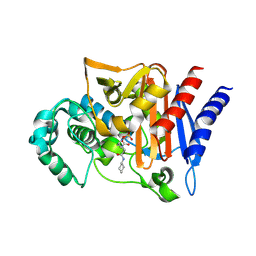 | | Amp-c beta-lactamase (pseudomonas aeruginosa) in complex with mk-7655 | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Scapin, G, Lu, J, Fitzgerald, P.M.D, Sharma, N. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of MK-7655, a beta-lactamase inhibitor for combination with Primaxin().
Bioorg.Med.Chem.Lett., 24, 2014
|
|
6D4V
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 22 (VCC061422) | | Descriptor: | 2-cyclohexyl-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6G91
 
 | |
4BCK
 
 | | Structure of CDK2 in complex with cyclin A and a 2-amino-4-heteroaryl- pyrimidine inhibitor | | Descriptor: | 3-[[5-cyano-4-[4-methyl-2-(methylamino)-1,3-thiazol-5-yl]pyrimidin-2-yl]amino]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2, ... | | Authors: | Hole, A.J, Baumli, S, Wang, S, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2012-10-02 | | Release date: | 2013-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Comparative Structural and Functional Studies of 4-(Thiazol- 5-Yl)-2-(Phenylamino)Pyrimidine-5-Carbonitrile Cdk9 Inhibitors Suggest the Basis for Isotype Selectivity.
J.Med.Chem., 56, 2013
|
|
6G9K
 
 | |
6D4R
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 18 (VCC399134) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, hydroxy(3-{4-[(isoquinolin-5-yl)sulfonyl]piperazine-1-carbonyl}phenyl)oxoammonium | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
6D5Y
 
 | | Crystal structure of ERK2 G169D mutant | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Yin, J, Jaiswal, B.S, Wang, W. | | Deposit date: | 2018-04-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | ERK Mutations and Amplification Confer Resistance to ERK-Inhibitor Therapy.
Clin. Cancer Res., 24, 2018
|
|
6D6U
 
 | | Human GABA-A receptor alpha1-beta2-gamma2 subtype in complex with GABA and flumazenil, conformation A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Noviello, C.M, Teng, J, Walsh Jr, R.M, Kim, J.J, Hibbs, R.E. | | Deposit date: | 2018-04-22 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structure of a human synaptic GABAAreceptor.
Nature, 559, 2018
|
|
4NFR
 
 | |
6GE0
 
 | |
6D4U
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor Compound 27 (VCC663664) | | Descriptor: | 2-(2,4-dimethoxyphenyl)-1-{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}ethan-1-one, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase | | Authors: | Ascher, D.B, Pacitto, A, Blundell, T.L. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH.
Eur.J.Med.Chem., 174, 2019
|
|
7E5M
 
 | | crystal structure of trans assembled human TROP-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor-associated calcium signal transducer 2 | | Authors: | Sun, M, Zhang, H, Chai, Y, Qi, J, Gao, G.F, Tan, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the cis and trans assembly of human trophoblast cell surface antigen 2.
Iscience, 24, 2021
|
|
7RXZ
 
 | | human Hsp90_MC domain structure | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.152 Å) | | Cite: | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
7RY0
 
 | | human Hsp90_MC domain structure | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
4A23
 
 | | Mus musculus Acetylcholinesterase in complex with racemic C5685 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Andersson, C.D, Artrursson, E, Hornberg, A, Tunemalm, A.K, Linusson, A, Ekstrom, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling.
Plos One, 6, 2011
|
|
