1MZP
 
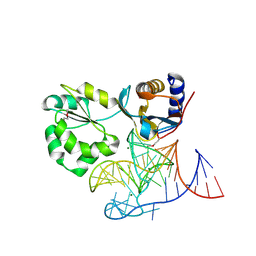 | | Structure of the L1 protuberance in the ribosome | | Descriptor: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | Authors: | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | Deposit date: | 2002-10-09 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
6LKN
 
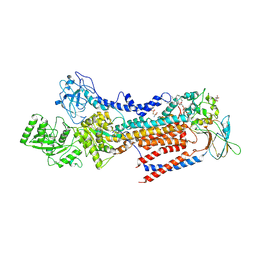 | | Crystal structure of ATP11C-CDC50A in PtdSer-bound E2P state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Hasegawa, K. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of a human plasma membrane phospholipid flippase.
J.Biol.Chem., 295, 2020
|
|
4GLL
 
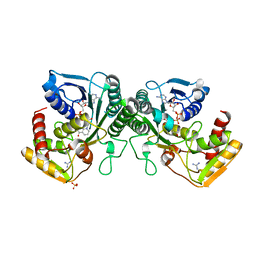 | | Crystal structure of human UDP-xylose synthase. | | Descriptor: | GUANIDINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Wood, Z.A, Polizzi, S.J. | | Deposit date: | 2012-08-14 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human UDP-alpha-d-xylose Synthase and Escherichia coli ArnA Conserve a Conformational Shunt That Controls Whether Xylose or 4-Keto-Xylose Is Produced.
Biochemistry, 51, 2012
|
|
1MTZ
 
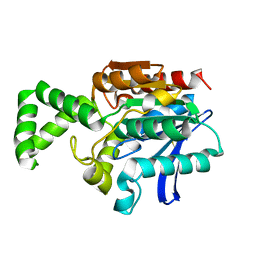 | | Crystal Structure of the Tricorn Interacting Factor F1 | | Descriptor: | Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
5W9J
 
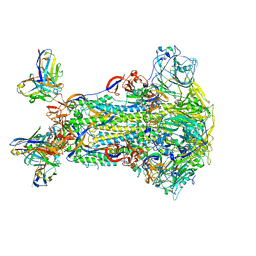 | |
1N0W
 
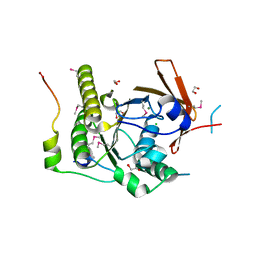 | | Crystal structure of a RAD51-BRCA2 BRC repeat complex | | Descriptor: | 1,2-ETHANEDIOL, ARTIFICIAL GLY-SER-MSE-GLY PEPTIDE, Breast cancer type 2 susceptibility protein, ... | | Authors: | Pellegrini, L, Yu, D.S, Lo, T, Anand, S, Lee, M, Blundell, T.L, Venkitaraman, A.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into DNA recombination from the structure of a RAD51-BRCA2 complex
Nature, 420, 2002
|
|
5WTT
 
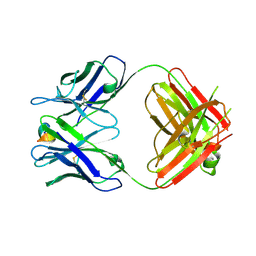 | | Structure of the 093G9 Fab in complex with the epitope peptide | | Descriptor: | Epitope peptide of Cyr61, Heavy chain of 093G9 Fab, Light chain of 093G9 Fab | | Authors: | Zhong, C, Hu, K, Shen, J, Ding, J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the recognition of CCN1 by monoclonal antibody 093G9.
J. Mol. Recognit., 30, 2017
|
|
1N1V
 
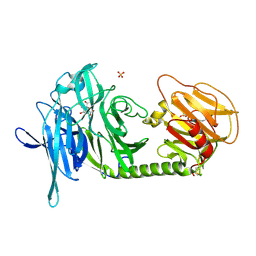 | | Trypanosoma rangeli sialidase in complex with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, SULFATE ION, Sialidase | | Authors: | Amaya, M.F, Buschiazzo, A, Nguyen, T, Alzari, P.M. | | Deposit date: | 2002-10-21 | | Release date: | 2003-01-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high resolution structures of free and
inhibitor-bound Trypanosoma rangeli
sialidase and its comparison with T.
cruzi trans-sialidase
J.Mol.Biol., 325, 2003
|
|
5WQX
 
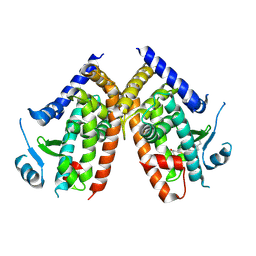 | | Covalent bond formation of synthetic ligand with hPPARg-LBD | | Descriptor: | 2-[E-(E-2-oxidanylidenehexadec-5-enylidene)amino]ethanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Kojima, H, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-11-29 | | Release date: | 2017-11-22 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | On-site reaction for PPAR gamma modification using a specific bifunctional ligand
Bioorg. Med. Chem., 25, 2017
|
|
5W9P
 
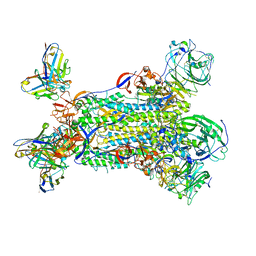 | |
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1MTQ
 
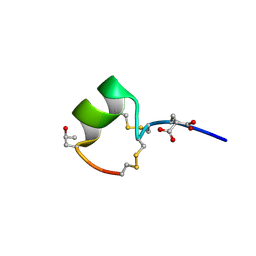 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID BY NMR SPECTROSCOPY | | Descriptor: | alpha-conotoxin GID | | Authors: | Nicke, A, Loughnan, M.L, Millard, E.L, Alewood, P.F, Adams, D.J, Daly, N.L, Craik, D.J, Lewis, R.J. | | Deposit date: | 2002-09-22 | | Release date: | 2003-02-11 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Isolation, Structure, and Activity of GID, a Novel alpha 4/7-Conotoxin with an Extended N-terminal Sequence
J.BIOL.CHEM., 278, 2003
|
|
2G7J
 
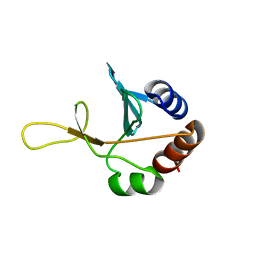 | | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72. | | Descriptor: | putative cytoplasmic protein | | Authors: | Aramini, J.M, Swapna, G.V.T, Ramelot, T.A, Ho, C.K, Cunningham, K, Ma, L.-C, Shetty, K, Xiao, R, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-02-28 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the putative cytoplasmic protein ygaC from Salmonella typhimurium. Northeast Structural Genomics target StR72.
To be Published
|
|
1MU0
 
 | | Crystal Structure of the Tricorn Interacting Factor F1 Complex with PCK | | Descriptor: | (2R,3S)-3-AMINO-1-CHLORO-4-PHENYL-BUTAN-2-OL, Proline iminopeptidase | | Authors: | Goettig, P, Groll, M, Kim, J.-S, Huber, R, Brandstetter, H. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the tricorn-interacting aminopeptidase F1 with different ligands explain its catalytic mechanism
Embo J., 21, 2002
|
|
7SCG
 
 | | FH210 bound Mu Opioid Receptor-Gi Protein Complex | | Descriptor: | (2E)-N-[(2S)-2-(dimethylamino)-3-(4-hydroxyphenyl)propyl]-3-(naphthalen-1-yl)prop-2-enamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, H, Kobilka, B. | | Deposit date: | 2021-09-28 | | Release date: | 2022-04-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-Based Evolution of G Protein-Biased mu-Opioid Receptor Agonists.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1MZJ
 
 | | Crystal Structure of the Priming beta-Ketosynthase from the R1128 Polyketide Biosynthetic Pathway | | Descriptor: | ACETYL GROUP, Beta-ketoacylsynthase III, COENZYME A | | Authors: | Pan, H, Tsai, S.C, Meadows, E.S, Miercke, L.J.W, Keatinge-Clay, A, O'Connell, J, Khosla, C, Stroud, R.M. | | Deposit date: | 2002-10-08 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the priming beta-ketosynthase from the
R1128 polyketide biosynthetic pathway
Structure, 10
|
|
7D9O
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Compound 2 | | Descriptor: | (2R)-2-[[4-fluoranyl-1-[(4-fluorophenyl)methyl]piperidin-4-yl]methyl]-5,6-dimethoxy-2,3-dihydroinden-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Liu, Q.F, Yin, W.C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
5W9M
 
 | |
1MZS
 
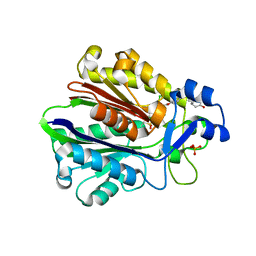 | | CRYSTAL STRUCTURE OF BETA-KETOACYL-ACP SYNTHASE III WITH BOUND dichlorobenzyloxy-indole-carboxylic acid inhibitor | | Descriptor: | 1-(5-CARBOXYPENTYL)-5-(2,6-DICHLOROBENZYLOXY)-1H-INDOLE-2-CARBOXYLIC ACID, 3-oxoacyl-[acyl-carrier-protein] synthase III, PHOSPHATE ION | | Authors: | Daines, R.A, Pendrak, I, Sham, K, Van Aller, G.S, Konstantinidis, A.K, Lonsdale, J.T, Janson, C.A, Qui, X, Brandt, M, Silverman, C, Head, M.S. | | Deposit date: | 2002-10-09 | | Release date: | 2002-11-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | First X-ray cocrystal structure of a bacterial FabH condensing enzyme and a small molecule inhibitor achieved using rational design and homology modeling
J.Med.Chem., 46, 2003
|
|
1N3N
 
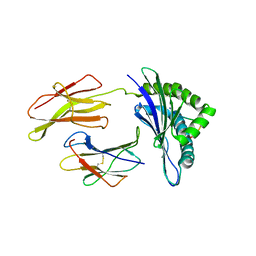 | | Crystal structure of a mycobacterial hsp60 epitope with the murine class I MHC molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Capitani, G, Tissot, A.C, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2002-10-29 | | Release date: | 2003-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of mycobacterial
and murine hsp60 epitopes in
complex with the class I MHC
molecule H-2D(b)
FEBS Lett., 543, 2003
|
|
5WUX
 
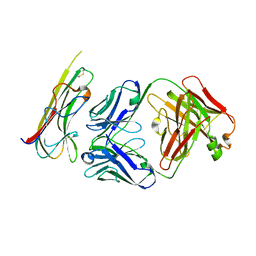 | | TNFalpha-certolizumab Fab | | Descriptor: | Tumor necrosis factor alpha, heavy, light | | Authors: | Heo, Y.S, Lee, J.U. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Int J Mol Sci, 18, 2017
|
|
5W9O
 
 | |
3VHU
 
 | |
3VZW
 
 | |
4GO0
 
 | |
