4XTW
 
 | | Mycobacterium tuberculosis biotin ligase complexed with bisubstrate inhibitor 46 with azide in place of 2'OH | | Descriptor: | 1,2-ETHANEDIOL, 2'-azido-2',5'-dideoxy-5'-[({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}sulfamoyl)amino]adenosine, Bifunctional ligase/repressor BirA | | Authors: | De la Mora-Rey, T, Finzel, B.C. | | Deposit date: | 2015-01-24 | | Release date: | 2015-09-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.30014467 Å) | | Cite: | Targeting Mycobacterium tuberculosis Biotin Protein Ligase (MtBPL) with Nucleoside-Based Bisubstrate Adenylation Inhibitors.
J.Med.Chem., 58, 2015
|
|
4XTZ
 
 | |
6PE4
 
 | | Yeast Vo motor in complex with 1 VopQ molecule | | Descriptor: | Cation transporter, Uncharacterized protein YPR170W-B, V-type proton ATPase subunit a, ... | | Authors: | Peng, W, Li, Y, Tomchick, D.R, Orth, K. | | Deposit date: | 2019-06-20 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A distinct inhibitory mechanism of the V-ATPase by Vibrio VopQ revealed by cryo-EM.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6P78
 
 | | queuine lyase from Clostridium spiroforme bound to SAM and queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5S)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-1,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, IRON/SULFUR CLUSTER, Queuine lyase, ... | | Authors: | Almo, S.C, Grove, T.L. | | Deposit date: | 2019-06-05 | | Release date: | 2019-09-18 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | Discovery of novel bacterial queuine salvage enzymes and pathways in human pathogens.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PF4
 
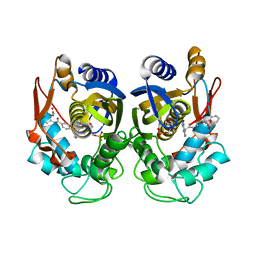 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)phenyl)acetic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)phenyl]acetic acid | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6FB9
 
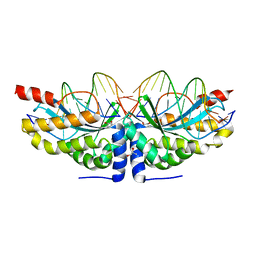 | |
6P7L
 
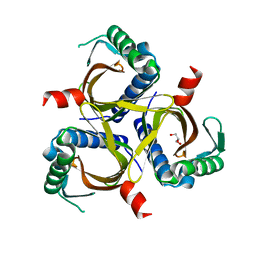 | |
6PFE
 
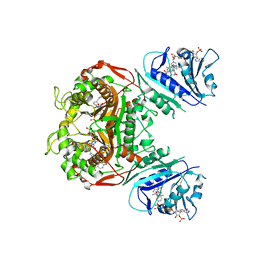 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxybenzoic acid. | | Descriptor: | 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxybenzoic acid, 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.812 Å) | | Cite: | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
6P7W
 
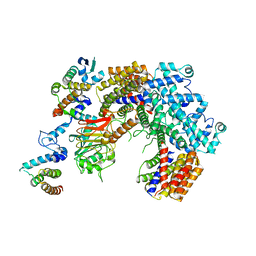 | | Structure of the K. lactis CBF3 core - Ndc10 D1 complex | | Descriptor: | Cep3, Ctf13, Ndc10, ... | | Authors: | Lee, P.D, Wei, H, Tan, D, Harrison, S.C. | | Deposit date: | 2019-06-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the Centromere Binding Factor 3 Complex from Kluyveromyces lactis.
J.Mol.Biol., 431, 2019
|
|
4XUL
 
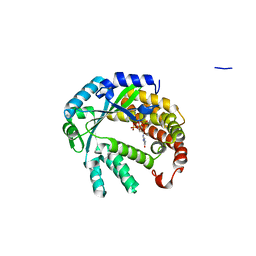 | | Crystal structure of M. chilensis Mg662 protein complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Lartigue, A, Priet, S, Claverie, J.M, Abergel, C. | | Deposit date: | 2015-01-26 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of M. chilensis Mg662 protein complexed with GTP
To Be Published
|
|
6P8H
 
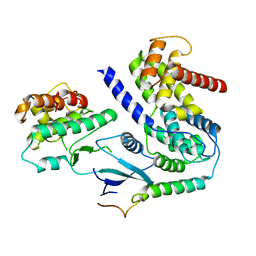 | | Crystal structure of CDK4 in complex with CyclinD1 and P21 | | Descriptor: | Cyclin-dependent kinase 4, Cyclin-dependent kinase inhibitor 1, G1/S-specific cyclin-D1 | | Authors: | Guiley, K.Z, Stevenson, J.W, Lou, K, Barkovich, K.J, Bunch, K, Tripathi, S.M, Shokat, K.M, Rubin, S.M. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition.
Science, 366, 2019
|
|
6PFZ
 
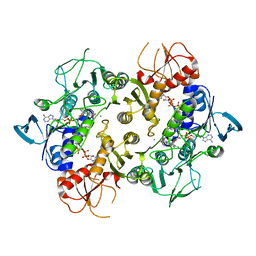 | | Structure of a NAD-Dependent Persulfide Reductase from A. fulgidus | | Descriptor: | CALCIUM ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sazinsky, M.H, Shabdar, S, Garcia-Constineiras, A, Crane III, E.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.10003877 Å) | | Cite: | Structural and Kinetic Characterization of Hyperthermophilic NADH-Dependent Persulfide Reductase from Archaeoglobus fulgidus .
Archaea, 2021, 2021
|
|
6P8P
 
 | | Structure of P. aeruginosa ATCC27853 HORMA1 | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-06-07 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.635 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
4XUZ
 
 | | Structure of CTX-M-15 bound to RPX-7009 at 1.5 A | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Clifton, M.C, Gardberg, A. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of a Cyclic Boronic Acid beta-Lactamase Inhibitor (RPX7009) with Utility vs Class A Serine Carbapenemases.
J.Med.Chem., 58, 2015
|
|
6EX9
 
 | |
6PHP
 
 | |
6OZ1
 
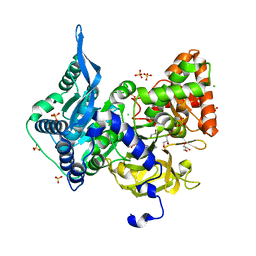 | | Crystal structure of the adenylation (A) domain of the carboxylate reductase (CAR) GR01_22995 from Mycobacterium chelonae | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Fedorchuk, T, Khusnutdinova, A, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | One-Pot Biocatalytic Transformation of Adipic Acid to 6-Aminocaproic Acid and 1,6-Hexamethylenediamine Using Carboxylic Acid Reductases and Transaminases.
J.Am.Chem.Soc., 142, 2020
|
|
6ESG
 
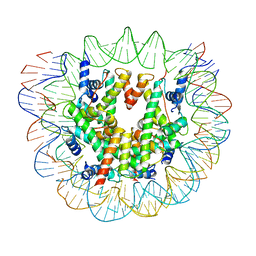 | | Nucleosome breathing : Class 2 | | Descriptor: | DNA (141-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Bilokapic, S, Halic, M. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Histone octamer rearranges to adapt to DNA unwrapping.
Nat. Struct. Mol. Biol., 25, 2018
|
|
2KQE
 
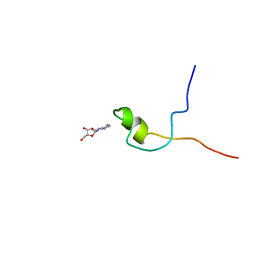 | | Second PBZ domain of human APLF protein in complex with ribofuranosyladenosine | | Descriptor: | ADENOSINE, Aprataxin and PNK-like factor, ZINC ION, ... | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
6PIW
 
 | |
6EU1
 
 | | RNA Polymerase III - open DNA complex (OC-POL3). | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
6PJ1
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-4(AJ-74) | | Descriptor: | 1,2-ETHANEDIOL, 1-methylcyclopentyl [(2R,6S,12Z,13aS,14aR,16aS)-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-14a-{[(1-methylcyclopropyl)sulfonyl]carbamoyl}-5,16-dioxo-1,2,3,5,6,7,8,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-6-yl]carbamate, NS3/4A protease, ... | | Authors: | Zephyr, J, Schiffer, C.A. | | Deposit date: | 2019-06-27 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Avoiding Drug Resistance by Substrate Envelope-Guided Design: Toward Potent and Robust HCV NS3/4A Protease Inhibitors.
Mbio, 11, 2020
|
|
6F0H
 
 | | Crystal structure ASF1-ip4 | | Descriptor: | CITRIC ACID, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
6PJL
 
 | | HIV-1 Protease NL4-3 WT in Complex with LR3-95 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(2S,4S,5S)-4-hydroxy-5-{[N-(methoxycarbonyl)-L-alloisoleucyl]amino}-1,6-diphenylhexan-2-yl]carbamate, Protease NL4-3, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | Structural Analysis of Potent Hybrid HIV-1 Protease Inhibitors Containing Bis-tetrahydrofuran in a Pseudosymmetric Dipeptide Isostere.
J.Med.Chem., 63, 2020
|
|
6EVM
 
 | | Crystal structure of a Pro-9 complexed peptide-substrate-binding domain of human type II collagen prolyl 4-hydroxylase | | Descriptor: | DIMETHYL SULFOXIDE, Pro-9, Prolyl 4-hydroxylase subunit alpha-2, ... | | Authors: | Murthy, A.V, Sulu, R, Koski, M.K, Wierenga, R.K. | | Deposit date: | 2017-11-02 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
Protein Sci., 27, 2018
|
|
