1U7O
 
 | | Magnesium Dependent Phosphatase 1 (MDP-1) | | Descriptor: | ACETATE ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily
Biochemistry, 43, 2004
|
|
3RT1
 
 | |
3N5F
 
 | | Crystal Structure of L-N-carbamoylase from Geobacillus stearothermophilus CECT43 | | Descriptor: | CACODYLATE ION, COBALT (II) ION, ISOPROPYL ALCOHOL, ... | | Authors: | Garcia-Pino, A, Martinez-Rodriguez, S, Gavira, J.A. | | Deposit date: | 2010-05-25 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Mutational and structural analysis of L-N-carbamoylase reveals new insights into a peptidase m20/m25/m40 family member.
J.Bacteriol., 194, 2012
|
|
4XSS
 
 | | Insulin-like growth factor I in complex with site 1 of a hybrid insulin receptor / Type 1 insulin-like growth factor receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor, ... | | Authors: | Lawrence, C, Kong, G.K.-W, Menting, J.G, Lawrence, M.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Congruency of Ligand Binding to the Insulin and Insulin/Type 1 Insulin-like Growth Factor Hybrid Receptors.
Structure, 23, 2015
|
|
5XVN
 
 | | E. far Cas1-Cas2/prespacer binary complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Xiao, Y, Ng, S, Nam, K.H, Ke, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration.
Nature, 550, 2017
|
|
4Y5Y
 
 | | Diabody 330 complex with EpoR | | Descriptor: | Erythropoietin receptor, GLYCEROL, diabody 330 VH domain, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
3N8I
 
 | |
3FAK
 
 | |
1YIC
 
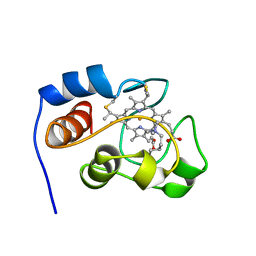 | | THE OXIDIZED SACCHAROMYCES CEREVISIAE ISO-1-CYTOCHROME C, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C, ISO-1, HEME C | | Authors: | Banci, L, Bertini, I, Bren, K.L, Gray, H.B, Sompornpisut, P, Turano, P. | | Deposit date: | 1997-02-18 | | Release date: | 1997-07-23 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of oxidized Saccharomyces cerevisiae iso-1-cytochrome c.
Biochemistry, 36, 1997
|
|
4KC3
 
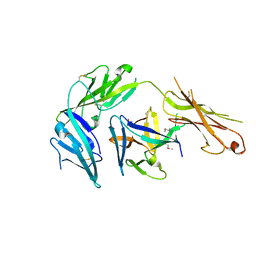 | | Cytokine/receptor binary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor-like 1, Interleukin-33 | | Authors: | Liu, X, Wang, X.Q. | | Deposit date: | 2013-04-24 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2702 Å) | | Cite: | Structural insights into the interaction of IL-33 with its receptors.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3F1N
 
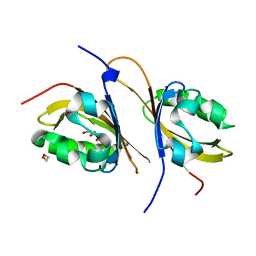 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1EE2
 
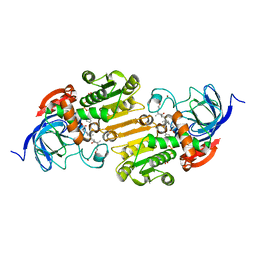 | |
3ICB
 
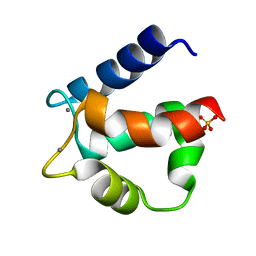 | | THE REFINED STRUCTURE OF VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN FROM BOVINE INTESTINE. MOLECULAR DETAILS, ION BINDING, AND IMPLICATIONS FOR THE STRUCTURE OF OTHER CALCIUM-BINDING PROTEINS | | Descriptor: | CALCIUM ION, CALCIUM-BINDING PROTEIN, SULFATE ION | | Authors: | Szebenyi, D.M.E, Moffat, K. | | Deposit date: | 1986-09-09 | | Release date: | 1986-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The refined structure of vitamin D-dependent calcium-binding protein from bovine intestine. Molecular details, ion binding, and implications for the structure of other calcium-binding proteins.
J.Biol.Chem., 261, 1986
|
|
1PJL
 
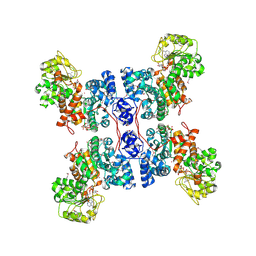 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
1QZC
 
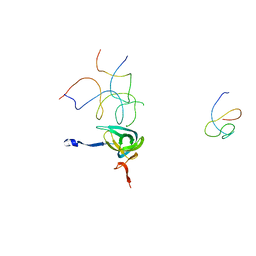 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZD
 
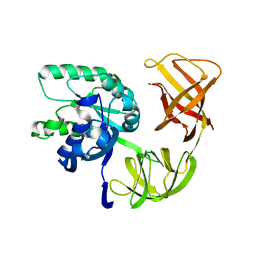 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
7YMB
 
 | |
1HYN
 
 | |
1QZA
 
 | | Coordinates of the A/T site tRNA model fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
2R4F
 
 | | Substituted Pyrazoles as Hepatselective HMG-COA reductase inhibitors | | Descriptor: | (3R,5R)-7-[1-(4-fluorophenyl)-4-(1-methylethyl)-3-{methyl[(1R)-1-phenylethyl]carbamoyl}-1H-pyrazol-5-yl]-3,5-dihydroxyheptanoic acid, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, SULFATE ION | | Authors: | Pavlovsky, A, Pfefferkorn, J.A, Harris, M.S, Finzel, B.C. | | Deposit date: | 2007-08-31 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substituted pyrazoles as hepatoselective HMG-CoA reductase inhibitors: discovery of (3R,5R)-7-[2-(4-fluoro-phenyl)-4-isopropyl-5-(4-methyl-benzylcarbamoyl)-2H-pyrazol-3-yl]-3,5-dihydroxyheptanoic acid (PF-3052334) as a candidate for the treatment of hypercholesterolemia.
J.Med.Chem., 51, 2008
|
|
8POM
 
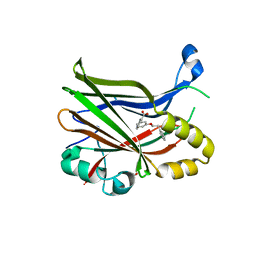 | | TEAD2 in complex with an inhibitor | | Descriptor: | 2-[[3-(2-phenylethoxy)phenyl]amino]pyridine-3-carboxylic acid, GLYCEROL, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
7PTQ
 
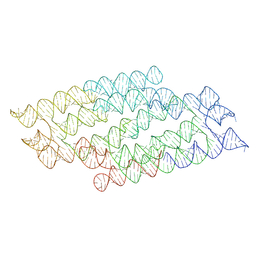 | | RNA origami 5-helix tile | | Descriptor: | Chains: C | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
8POJ
 
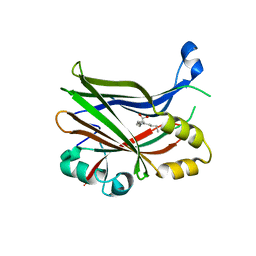 | | TEAD2 in complex with an inhibitor | | Descriptor: | 4-fluoranyl-2-[(3-phenylphenyl)amino]benzoic acid, MYRISTIC ACID, Transcriptional enhancer factor TEF-4 | | Authors: | Guichou, J.F. | | Deposit date: | 2023-07-05 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development of LM-41 and AF-2112, two flufenamic acid-derived TEAD inhibitors obtained through the replacement of the trifluoromethyl group by aryl rings.
Bioorg.Med.Chem.Lett., 95, 2023
|
|
7PTS
 
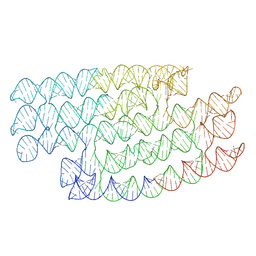 | | RNA origami 5-helix tile | | Descriptor: | 5HT-B | | Authors: | McRae, E.K.S, Andersen, E.S. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.71 Å) | | Cite: | Structure, folding and flexibility of co-transcriptional RNA origami.
Nat Nanotechnol, 18, 2023
|
|
7PTK
 
 | |
