8TDD
 
 | | Structure of PYCR1 complexed with NADH and 2-(furan-2-yl)acetic acid | | Descriptor: | (furan-2-yl)acetic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
8TD8
 
 | | Structure of PYCR1 complexed with NADH and 2S-Hydroxy-3,3-dimethylbutyric acid | | Descriptor: | (2S)-2-hydroxy-3,3-dimethylbutanoic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
8TD6
 
 | | Structure of PYCR1 complexed with NADH and 2-(Methylthio)acetic acid | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, mitochondrial, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
8TD3
 
 | |
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
8TDB
 
 | | Structure of PYCR1 complexed with NADH and 1-hydroxyethane-1-sulfonate | | Descriptor: | (1R)-1-hydroxyethane-1-sulfonic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Pyrroline-5-carboxylate reductase 1, ... | | Authors: | Tanner, J.J, Meeks, K.R. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Screening a knowledge-based library of low molecular weight compounds against the proline biosynthetic enzyme 1-pyrroline-5-carboxylate 1 (PYCR1)
Protein Sci., 33, 2024
|
|
1EL4
 
 | | STRUCTURE OF THE CALCIUM-REGULATED PHOTOPROTEIN OBELIN DETERMINED BY SULFUR SAS | | Descriptor: | C2-HYDROXY-COELENTERAZINE, CHLORIDE ION, OBELIN | | Authors: | Liu, Z.J, Vysotski, E.S, Rose, J, Lee, J, Wang, B.C. | | Deposit date: | 2000-03-13 | | Release date: | 2001-03-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure of the Ca2+-regulated photoprotein obelin at 1.7 A resolution determined directly from its sulfur substructure.
Protein Sci., 9, 2000
|
|
8TD5
 
 | |
2XN9
 
 | | Crystal structure of the ternary complex between human T cell receptor, staphylococcal enterotoxin H and human major histocompatibility complex class II | | Descriptor: | ENTEROTOXIN H, GLYCEROL, HEMAGGLUTININ, ... | | Authors: | Saline, M, Rodstrom, K.E.J, Fischer, G, Orekhov, V.Y, Karlsson, B.G, Lindkvist-Petersson, K. | | Deposit date: | 2010-07-31 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of Superantigen Complexed with Tcr and Mhc Reveals Novel Insights Into Superantigenic T Cell Activation.
Nat.Commun., 1, 2010
|
|
1E0O
 
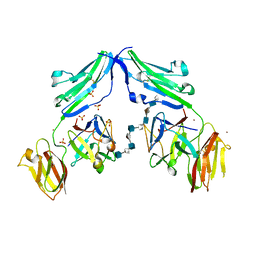 | | CRYSTAL STRUCTURE OF A TERNARY FGF1-FGFR2-HEPARIN COMPLEX | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, FIBROBLAST GROWTH FACTOR 1, FIBROBLAST GROWTH FACTOR RECEPTOR 2, ... | | Authors: | Pellegrini, L, Burke, D.F, von Delft, F, Mulloy, B, Blundell, T.L. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Fibroblast Growth Factor Receptor Ectodomain Bound to Ligand and Heparin
Nature, 407, 2000
|
|
1IHN
 
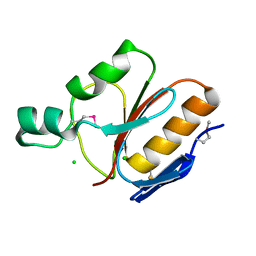 | |
4Y5V
 
 | | Diabody 305 complex with EpoR | | Descriptor: | DI(HYDROXYETHYL)ETHER, Diabody 305 VL domain, Erythropoietin receptor, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-04-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
3LJ9
 
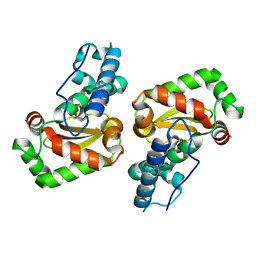 | | X-ray structure of the iron superoxide dismutase from pseudoalteromonas haloplanktis in complex with sodium azide | | Descriptor: | AZIDE ION, FE (III) ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, ... | | Authors: | Merlino, A, Russo Krauss, I, Rossi, B, Conte, M, Vergara, A, Sica, F. | | Deposit date: | 2010-01-26 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and flexibility in cold-adapted iron superoxide dismutases: the case of the enzyme isolated from Pseudoalteromonas haloplanktis.
J.Struct.Biol., 172, 2010
|
|
1QOS
 
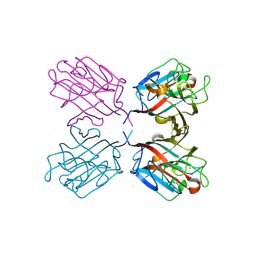 | | lectin UEA-II complexed with chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Loris, R, De Greve, H, Dao-Thi, M.-H, Messens, J, Imberty, A, Wyns, L. | | Deposit date: | 1999-11-16 | | Release date: | 2000-02-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by Lectin II from Ulex Europaeus, a Protein with a Promiscuous Carbohydrate Binding Site
J.Mol.Biol., 301, 2000
|
|
8FO3
 
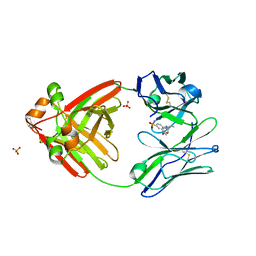 | |
5VST
 
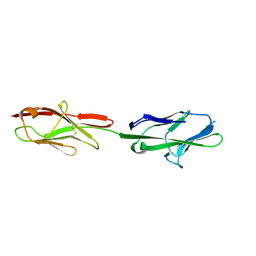 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
8FO4
 
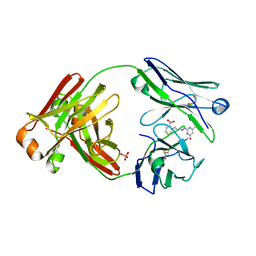 | |
8FO5
 
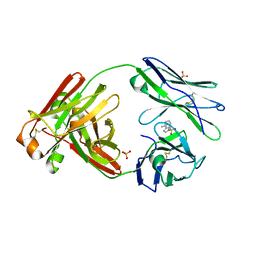 | |
1QLU
 
 | |
5HQY
 
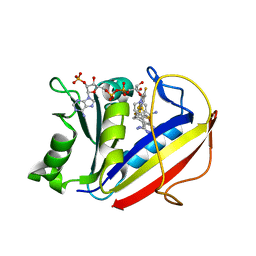 | | human dihydrofolate reductase complex with NADPH and 5-methyl-6-(2',3',4'-trifluorophenylthio)thieno[2,3-d]pyrimidine-2,4-diamine | | Descriptor: | 5-methyl-6-[(2,3,4-trifluorophenyl)sulfanyl]thieno[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2016-01-22 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Human dihydrofolate reductase ternary complex with a series of fluorine substituted 5-methyl-6-phenythio)thieno[2,3-d]pyrimidine-2,4-diamines
To Be Published
|
|
4Z9B
 
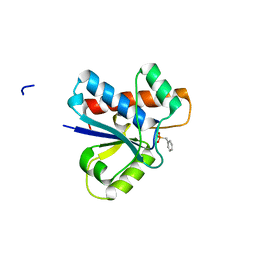 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with benzylphosphonic acid | | Descriptor: | Low molecular weight phosphotyrosine protein phosphatase, benzylphosphonic acid | | Authors: | Fonseca, E.M.B, Trivella, D.B.B, Scorsato, V, Dias, M.P, de Oliveira, F.L, Miranda, P.C.M.L, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
6QIN
 
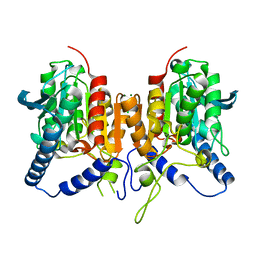 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PMGL2 | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-21 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
4Z9A
 
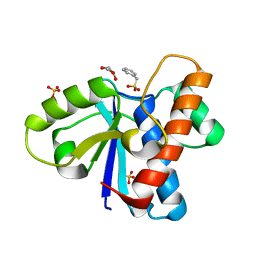 | | Crystal structure of Low Molecular Weight Protein Tyrosine Phosphatase isoform A complexed with phenylmethanesulfonic acid | | Descriptor: | GLYCEROL, Low molecular weight phosphotyrosine protein phosphatase, SULFATE ION, ... | | Authors: | Trivella, D.B.B, Fonseca, E.M.B, Scorsato, V, Dias, M.P, Aparicio, R. | | Deposit date: | 2015-04-10 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the apo form and a complex of human LMW-PTP with a phosphonic acid provide new evidence of a secondary site potentially related to the anchorage of natural substrates.
Bioorg.Med.Chem., 23, 2015
|
|
1HCW
 
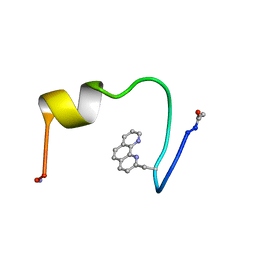 | |
6QLA
 
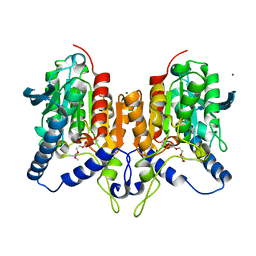 | | CRYSTAL STRUCTURE OF THE PMGL2 ESTERASE (point mutant 1) FROM PERMAFROST METAGENOMIC LIBRARY | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Boyko, K.M, Garsia, D, Nikolaeva, A.Y, Korzhenevskiy, D.A, Kryukova, M.V, Petrovskaya, L.E, Novototskaya-Vlasova, K.A, Rivkina, E.M, Dolgikh, D.A, Kirpichnikov, M.P, Popov, V.O. | | Deposit date: | 2019-01-31 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of PMGL2 esterase from the hormone-sensitive lipase family with GCSAG motif around the catalytic serine.
Plos One, 15, 2020
|
|
