9S5G
 
 | | Temporizin-1 NMR analysis | | Descriptor: | Temporizin-1 | | Authors: | Falcigno, L, D'Auria, G, Galdiero, S, Falanga, A, Bellavita, R. | | Deposit date: | 2025-07-29 | | Release date: | 2025-09-17 | | Last modified: | 2025-10-08 | | Method: | SOLUTION NMR | | Cite: | Temporizin-1 Meets the Membranes: Probing Membrane Inser-Tion and Disruption Mechanisms.
Antibiotics, 14, 2025
|
|
7DD4
 
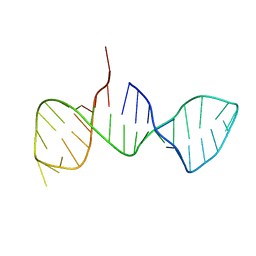 | |
6W1D
 
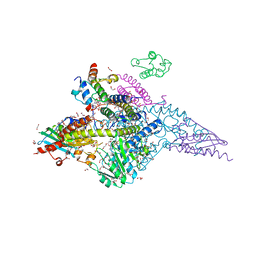 | | Structure of human mitochondrial complex Nfs1-ISCU2 (WT)-ISD11 with E.coli ACP1 at 1.8 A resolution (NIAU)2 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, ... | | Authors: | Boniecki, M.T, Cygler, M. | | Deposit date: | 2020-03-04 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
To Be Published
|
|
8SW6
 
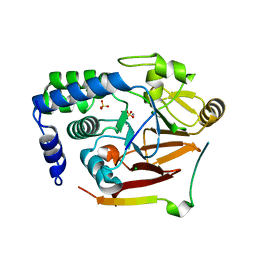 | | Protein Phosphatase 1 in complex with PP1-specific Phosphatase targeting peptide (PhosTAP) version 3 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PP1-specific Phosphatase-Targeting Peptide version 3, ... | | Authors: | Choy, M.S, Peti, W, Page, R. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A protein phosphatase 1 specific phos phatase ta rgeting p eptide (PhosTAP) to identify the PP1 phosphatome.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6W0Q
 
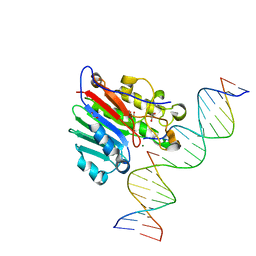 | | APE1 endonuclease product complex D148E | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Whitaker, A.M. | | Deposit date: | 2020-03-02 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Molecular and structural characterization of disease-associated APE1 polymorphisms.
DNA Repair (Amst.), 91-92, 2020
|
|
7OKP
 
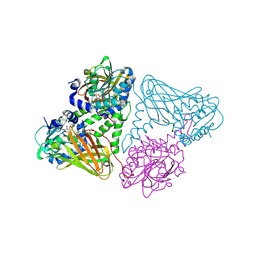 | | Crystal structure of mouse CARM1 in complex with histone H3_13-22 K18 acetylated | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.3, Histone-arginine methyltransferase CARM1, ... | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-05-18 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7OS4
 
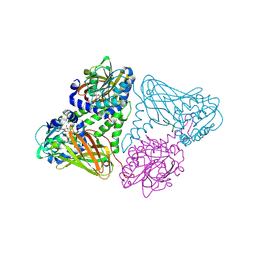 | | Crystal structure of mouse CARM1 in complex with histone H3_13-31 K18 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[(~{E})-prop-1-enyl]oxolane-3,4-diol, Histone H3.1, Histone-arginine methyltransferase CARM1 | | Authors: | Marechal, N, Cura, V, Troffer-Charlier, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Structural Studies Provide New Insights into the Role of Lysine Acetylation on Substrate Recognition by CARM1 and Inform the Design of Potent Peptidomimetic Inhibitors.
Chembiochem, 22, 2021
|
|
7MLX
 
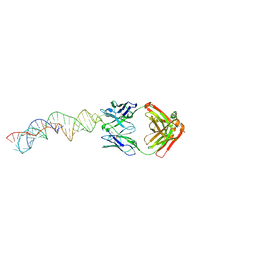 | |
8F11
 
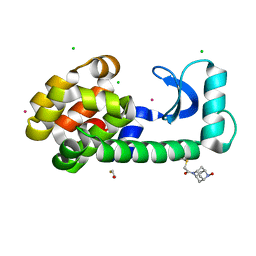 | | T4 lysozyme with a 2,6-diazaadamantane nitroxide (DZD) spin label | | Descriptor: | 1-[(1r,3r,5r,7r)-6-hydroxy-2,6-diazatricyclo[3.3.1.1~3,7~]decan-2-yl]ethan-1-one, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wilson, M.A, Madzelan, P, Rajca, A, Stein, R, Yang, Z. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Cucurbit[7]uril Enhances Distance Measurements of Spin-Labeled Proteins.
J.Am.Chem.Soc., 145, 2023
|
|
6W0Y
 
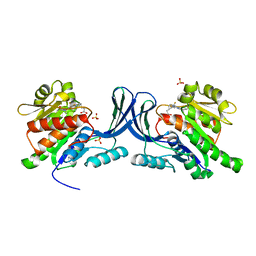 | |
7ALU
 
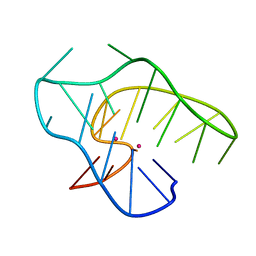 | |
7OCT
 
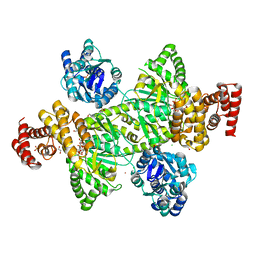 | | NADPH bound to the dehydrogenase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD-N374A from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8EPV
 
 | | 2.2 A crystal structure of the lipocalin cat allergen Fel d 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fel d 7 allergen, ... | | Authors: | Min, J, Pedersen, L.C, Geoffrey, M.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and ligand binding analysis of the pet allergens Can f 1 and Fel d 7.
Front Allergy, 4, 2023
|
|
6YKK
 
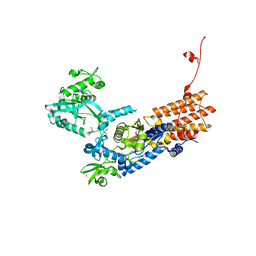 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
6O3F
 
 | |
6Y83
 
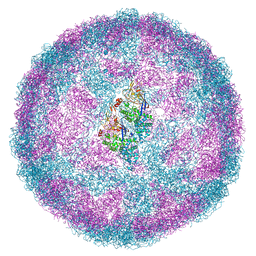 | | Capsid structure of Leishmania RNA virus 1 | | Descriptor: | Capsid protein | | Authors: | Prochazkova, M, Fuzik, T, Grybtchuk, D, Falginella, F, Podesvova, L, Yurchenko, V, Vacha, R, Plevka, P. | | Deposit date: | 2020-03-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Capsid Structure of Leishmania RNA Virus 1.
J.Virol., 95, 2021
|
|
8KB7
 
 | | Crystal structure of UDP/mannose-bound AGO61/beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Satoh, T, Umezawa, F, Yagi, H, Kato, K. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of UDP/mannose-bound AGO61/beta-1,4-N-Acetylglucosaminyltransferase 2 (POMGNT2)
To Be Published
|
|
6NNH
 
 | | Structure of Closed state of Dihydrofolate reductase from Mycobacterium tuberculosis in complex with NADPH and cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, COBALT (II) ION, Dihydrofolate reductase, ... | | Authors: | Giudice, J.H.P, Ribeiro, J.A, Dias, M.V.B. | | Deposit date: | 2019-01-15 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Crystal structures of the closed form of Mycobacterium tuberculosis dihydrofolate reductase in complex with dihydrofolate and antifolates.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8EQG
 
 | |
8EO4
 
 | |
8EQK
 
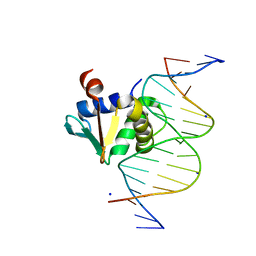 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAACCGGAAGTGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*CP*CP*GP*GP*AP*AP*GP*TP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*AP*CP*TP*TP*CP*CP*GP*GP*TP*TP*AP*T)-3'), SODIUM ION, ... | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-10-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
8EQL
 
 | |
8D1R
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT520 | | Descriptor: | 1-{3-[(4-chlorophenyl)methoxy]phenyl}methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
8D25
 
 | | Crystal structure of acetyltransferase Eis from Mycobacterium tuberculosis in complex with inhibitor SGT530 | | Descriptor: | 1-{2-[(3-chlorophenyl)methoxy]phenyl}-N-[(pyridin-4-yl)methyl]methanamine, GLYCEROL, N-acetyltransferase Eis | | Authors: | Pang, A.H, Punetha, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2022-05-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of substituted benzyloxy-benzylamine inhibitors of acetyltransferase Eis and their anti-mycobacterial activity.
Eur.J.Med.Chem., 242, 2022
|
|
8EO1
 
 | |
