7U3J
 
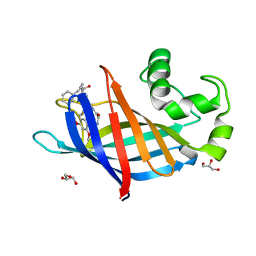 | | GID4 in complex with compound 88 | | Descriptor: | (2S)-2-{[(2S)-2-({N-[(2,4-dimethoxyphenyl)methyl]glycyl}amino)-2-(thiophen-2-yl)acetyl]amino}-N-methyl-4-phenylbutanamide, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
3QLY
 
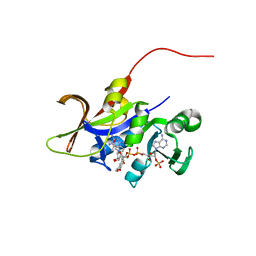 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Strain CBS138 chromosome J complete sequence | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
2G6U
 
 | |
7CKQ
 
 | |
2QUE
 
 | | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution | | Descriptor: | 4-METHOXYBENZOIC ACID, AJMALINE, Phospholipase A2 VRV-PL-VIIIa | | Authors: | Kumar, S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-08-05 | | Release date: | 2007-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Saturation of substrate-binding site using two natural ligands: Crystal structure of a ternary complex of phospholipase A2 with anisic acid and ajmaline at 2.25 A resolution
To be Published
|
|
1J16
 
 | | BENZAMIDINE IN COMPLEX WITH RAT TRYPSIN MUTANT X99/175/190RT | | Descriptor: | BENZAMIDINE, CALCIUM ION, SULFATE ION, ... | | Authors: | Stubbs, M.T. | | Deposit date: | 2002-11-30 | | Release date: | 2002-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reconstructing the Binding Site of Factor Xa in Trypsin Reveals Ligand-induced Structural Plasticity
J.MOL.BIOL., 325, 2003
|
|
2BAW
 
 | | Human Nuclear Receptor-Ligand Complex 1 | | Descriptor: | Peroxisome proliferator activated receptor delta, VACCENIC ACID, heptyl beta-D-glucopyranoside | | Authors: | Fyffe, S.A, Alphey, M.S, Buetow, L, Smith, T.K, Ferguson, M.A.J, Sorensen, M.D, Bjorkling, F, Hunter, W.N. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Reevaluation of the PPAR-beta/delta Ligand Binding Domain Model Reveals Why It Exhibits the Activated Form
Mol.Cell, 21, 2006
|
|
3WS6
 
 | | Crystal Structure of H-2D in complex with a mimotopic peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Kumar, P.R, Mukherjee, G, Samanta, D, DiLorenzo, T.P, Almo, S.C, Immune Function Network, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Compensatory mechanisms allow undersized anchor-deficient class I MHC ligands to mediate pathogenic autoreactive T cell responses
J. Immunol., 193, 2014
|
|
5PZM
 
 | |
3QLR
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
1P2E
 
 | | H61A mutant of flavocytochrome c3 | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella frigidimarina
Biochemistry, 42, 2003
|
|
1Y6L
 
 | | Human ubiquitin conjugating enzyme E2E2 | | Descriptor: | Ubiquitin-conjugating enzyme E2E2 | | Authors: | Walker, J.R, Avvakumov, G.V, Newman, E.M, Mackenzie, F, Kozieradzki, I, Bochkarev, A, Sundstrom, M, Arrowsmith, C, Edwards, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-06 | | Release date: | 2005-01-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3WS3
 
 | | Crystal Structure of H-2D in complex with an insulin derived peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Kumar, P.R, Mukherjee, G, Samanta, D, DiLorenzo, T.P, Almo, S.C, Immune Function Network, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-02-28 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.335 Å) | | Cite: | Compensatory mechanisms allow undersized anchor-deficient class I MHC ligands to mediate pathogenic autoreactive T cell responses
J. Immunol., 193, 2014
|
|
5JXJ
 
 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5U20
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni, internal PLP-aldimine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Thoden, J.B, Holden, H.M, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
5U23
 
 | | X-ray structure of the WlaRG aminotransferase from Campylobacter jejuni in complex with TDP-Qui3N | | Descriptor: | (2R,3R,4S,5S,6R)-3,5-dihydroxy-4-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}-6-methyltetrahydro-2H-pyran-2-yl [(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Holden, H.M, Thoden, J.B, Dow, G.T, Gilbert, M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.
Protein Sci., 26, 2017
|
|
4PUL
 
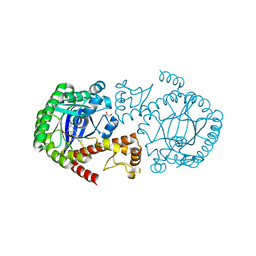 | | tRNA-Guanine Transglycosylase (TGT) Mutant D102N in Complex with 6-Amino-2-(methylamino)-1H,7H,8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-(methylamino)-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Neeb, M, Heine, A, Klebe, G. | | Deposit date: | 2014-03-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.654 Å) | | Cite: | Chasing Protons: How Isothermal Titration Calorimetry, Mutagenesis, and pKa Calculations Trace the Locus of Charge in Ligand Binding to a tRNA-Binding Enzyme.
J.Med.Chem., 57, 2014
|
|
4LPH
 
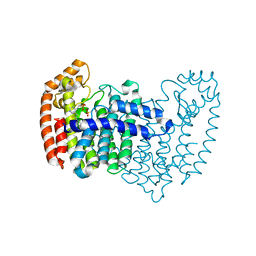 | | Crystal structure of human FPPS in complex with CL03093 | | Descriptor: | ({[6-(4-methylphenyl)thieno[2,3-d]pyrimidin-4-yl]amino}methyl)phosphonic acid, Farnesyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Park, J, Leung, C.Y, Tsantrizos, Y.S, Berghuis, A.M. | | Deposit date: | 2013-07-16 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multistage screening reveals chameleon ligands of the human farnesyl pyrophosphate synthase: implications to drug discovery for neurodegenerative diseases.
J.Med.Chem., 57, 2014
|
|
1D6M
 
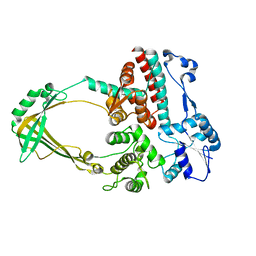 | |
2DJY
 
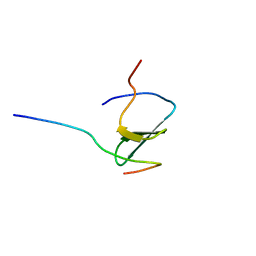 | | Solution structure of Smurf2 WW3 domain-Smad7 PY peptide complex | | Descriptor: | Mothers against decapentaplegic homolog 7, Smad ubiquitination regulatory factor 2 | | Authors: | Chong, P.A, Lin, H, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2006-04-06 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Expanded WW Domain Recognition Motif Revealed by the Interaction between Smad7 and the E3 Ubiquitin Ligase Smurf2.
J.Biol.Chem., 281, 2006
|
|
1UMB
 
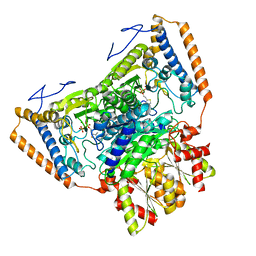 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
2ARA
 
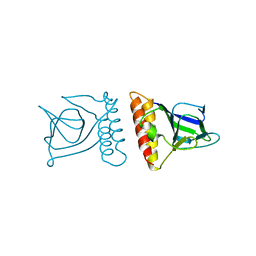 | |
6CK5
 
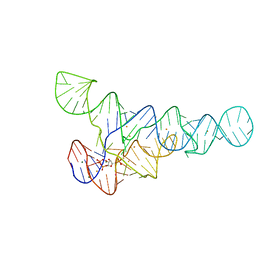 | | PRPP riboswitch from T. mathranii bound to PRPP | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, BARIUM ION, MAGNESIUM ION, ... | | Authors: | Knappenberger, A.J, Reiss, C.W, Strobel, S.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of two aptamers with differing ligand specificity reveal ruggedness in the functional landscape of RNA.
Elife, 7, 2018
|
|
4MNV
 
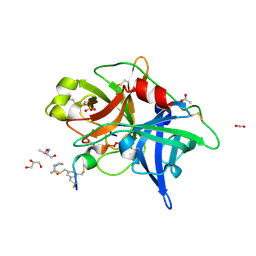 | | Crystal structure of bicyclic peptide UK729 bound as an acyl-enzyme intermediate to urokinase-type plasminogen activator (uPA) | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, ACETATE ION, GLYCEROL, ... | | Authors: | Chen, S, Pojer, F, Heinis, C. | | Deposit date: | 2013-09-11 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptide ligands stabilized by small molecules.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1QI2
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE TETRAGONAL CRYSTAL FORM IN COMPLEX WITH 2',4'-DINITROPHENYL 2-DEOXY-2-FLUORO-B-D-CELLOTRIOSIDE | | Descriptor: | CALCIUM ION, ENDOGLUCANASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-2-deoxy-2-fluoro-alpha-D-glucopyranose | | Authors: | Varrot, A, Schulein, M, Davies, G.J. | | Deposit date: | 1999-06-02 | | Release date: | 2000-06-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Insights into ligand-induced conformational change in Cel5A from Bacillus agaradhaerens revealed by a catalytically active crystal form.
J.Mol.Biol., 297, 2000
|
|
