7UL5
 
 | |
7UL3
 
 | | CryoEM Structure of Inactive H2R Bound to Famotidine, Nb6M, and NabFab | | Descriptor: | Histamine H2 receptor, NabFab HC, NabFab LC, ... | | Authors: | Robertson, M.J, Skiniotis, G. | | Deposit date: | 2022-04-03 | | Release date: | 2022-06-29 | | Last modified: | 2022-12-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure determination of inactive-state GPCRs with a universal nanobody.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6WHA
 
 | | HTR2A bound to 25-CN-NBOH in complex with a mini-Galpha-q protein, beta/gamma subunits and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 4-(2-{[(2-hydroxyphenyl)methyl]amino}ethyl)-2,5-dimethoxybenzonitrile, G subunit q (Gi2-mini-Gq chimeric), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Skiniotis, G, Roth, B, Kim, K, Panova, O. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6YZY
 
 | |
1G7G
 
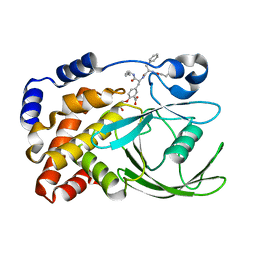 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXES WITH PNU179326 | | Descriptor: | 2-(CARBOXYMETHOXY)-5-[(2S)-2-({(2S)-2-[(3-CARBOXYPROPANOYL)AMINO] -3-PHENYLPROPANOYL}AMINO)-3-OXO-3-(PENTYLAMINO)PROPYL]BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Bleasdale, J.E, Ogg, D, Larsen, S.D. | | Deposit date: | 2000-11-10 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small molecule peptidomimetics containing a novel phosphotyrosine bioisostere inhibit protein tyrosine phosphatase 1B and augment insulin action.
Biochemistry, 40, 2001
|
|
8FPE
 
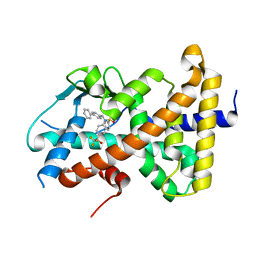 | | Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP | | Descriptor: | N-[([1,1'-biphenyl]-4-yl)methyl]-N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]benzenesulfonamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Huber, A.D, Poudel, S, Seetharaman, J, Miller, D.J, Lin, W, Li, Y, Chen, T. | | Deposit date: | 2023-01-04 | | Release date: | 2023-03-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided approach to modulate small molecule binding to a promiscuous ligand-activated protein.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3QSC
 
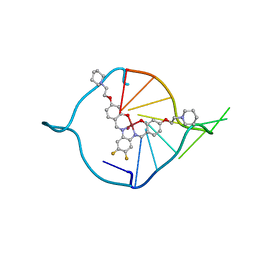 | | The first crystal structure of a human telomeric G-quadruplex DNA bound to a metal-containing ligand (a copper complex) | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*(BRU)P*AP*GP*GP*GP*TP*T)-3', POTASSIUM ION, [2,2'-{(4,5-difluorobenzene-1,2-diyl)bis[(nitrilo-kappaN)methylylidene]}bis{5-[2-(piperidin-1-yl)ethoxy]phenolato-kappaO}(2-)]copper (II) | | Authors: | Campbell, N.H, Abd Karim, N.H, Parkinson, G.N, Vilar, R, Neidle, S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of structure-activity relationships between salphen metal complexes and human telomeric DNA quadruplexes.
J.Med.Chem., 55, 2012
|
|
3QSF
 
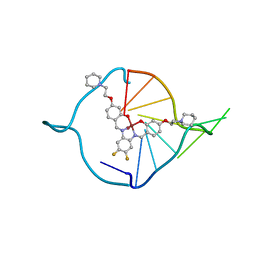 | | The first crystal structure of a human telomeric G-quadruplex DNA bound to a metal-containing ligand (a nickel complex) | | Descriptor: | 5'-D(*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*T)-3', POTASSIUM ION, [2,2'-{(4,5-difluorobenzene-1,2-diyl)bis[(nitrilo-kappaN)methylylidene]}bis{5-[2-(piperidin-1-yl)ethoxy]phenolato-kappa O}(2-)]nickel (II) | | Authors: | Campbell, N.H, Abd Karim, N.H, Parkinson, G.N, Vilar, R, Neidle, S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis of structure-activity relationships between salphen metal complexes and human telomeric DNA quadruplexes.
J.Med.Chem., 55, 2012
|
|
6QCC
 
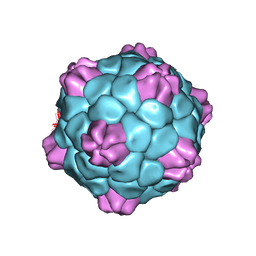 | | Cryo-EM Atomic Structure of Broad Bean Stain Virus (BBSV) | | Descriptor: | Large coat-protein subunit, Small coat-protein subunit | | Authors: | Lecorre, F, Lai Jee Him, J, Blanc, S, Zeddam, J.-L, Trapani, S, Bron, P. | | Deposit date: | 2018-12-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The cryo-electron microscopy structure of Broad Bean Stain Virus suggests a common capsid assembly mechanism among comoviruses.
Virology, 530, 2019
|
|
5JNN
 
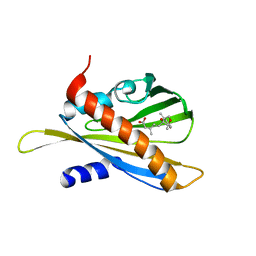 | | Crystal structure of abscisic acid receptor PYL2 in complex with Phaseic acid. | | Descriptor: | (3S,4E)-5-[(1R,5R,8S)-8-hydroxy-1,5-dimethyl-3-oxo-6-oxabicyclo[3.2.1]octan-8-yl]-3-methylpent-4-enoic acid, Abscisic acid receptor PYL2 | | Authors: | Weng, J.K, Noel, J.P. | | Deposit date: | 2016-04-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-evolution of Hormone Metabolism and Signaling Networks Expands Plant Adaptive Plasticity.
Cell, 166, 2016
|
|
8BHF
 
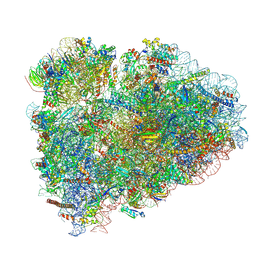 | | Cryo-EM structure of stalled rabbit 80S ribosomes in complex with human CCR4-NOT and CNOT4 | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S11, ... | | Authors: | Absmeier, E, Chandrasekaran, V, O'Reilly, F.J, Stowell, J.A.W, Rappsilber, J, Passmore, L.A. | | Deposit date: | 2022-10-31 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6WN4
 
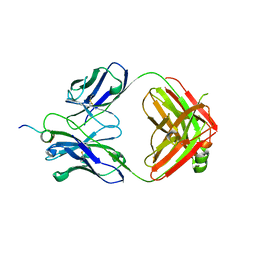 | | Structural basis for the binding of monoclonal antibody 5D2 to the tryptophan-rich lipid-binding loop in lipoprotein lipase | | Descriptor: | 5D2 FAB HEAVY CHAIN, 5D2 FAB LIGHT CHAIN, Lipoprotein lipase peptide | | Authors: | Luz, J.G, Birrane, G, Young, S.G, Meiyappan, M, Ploug, M. | | Deposit date: | 2020-04-22 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis for monoclonal antibody 5D2 binding to the tryptophan-rich loop of lipoprotein lipase.
J.Lipid Res., 61, 2020
|
|
7FZY
 
 | | Crystal Structure of human FABP4 in complex with 6-chloro-4-phenyl-2-piperidin-1-yl-3-(1H-tetrazol-5-yl)quinoline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-chloro-4-phenyl-2-(piperidin-1-yl)-3-(1H-tetrazol-5-yl)quinoline, DIMETHYL SULFOXIDE, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Kuhne, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
6Q61
 
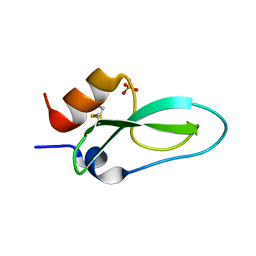 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | Kunitz-type conkunitzin-S1, SULFATE ION | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7RQQ
 
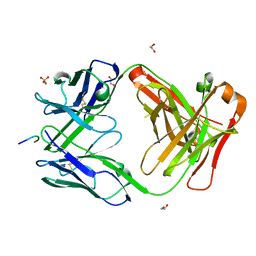 | |
7UMM
 
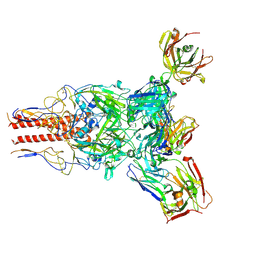 | | H1 Solomon Islands 2006 hemagglutinin in complex with Ab109 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Windsor, I.W, Caradonna, T.M, Schmidt, A.G. | | Deposit date: | 2022-04-07 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | An epitope-enriched immunogen expands responses to a conserved viral site.
Cell Rep, 41, 2022
|
|
7RQP
 
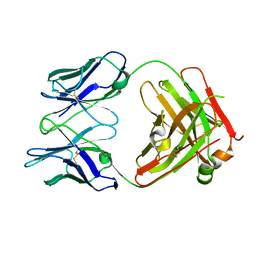 | | Structure of PfCSP NPNV binding antibody L9 | | Descriptor: | L9 Heavy Chain, L9 Kappa Chain | | Authors: | Hurlburt, N.K, Pancera, M. | | Deposit date: | 2021-08-06 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The light chain of the L9 antibody is critical for binding circumsporozoite protein minor repeats and preventing malaria.
Cell Rep, 38, 2022
|
|
7RQR
 
 | |
5J0Z
 
 | | Crystal structure of GLIC in complex with DHA | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Basak, S, Schmandt, N, Chakrapani, S. | | Deposit date: | 2016-03-28 | | Release date: | 2017-03-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure and dynamics of a lipid-induced potential desensitized-state of a pentameric ligand-gated channel.
Elife, 6, 2017
|
|
6VR4
 
 | |
6Q6C
 
 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | 1,2-ETHANEDIOL, Kunitz-type conkunitzin-S1, NITRATE ION, ... | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2HK3
 
 | |
8A9T
 
 | | Tubulin-[1,2]oxazoloisoindole-1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
6YA6
 
 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6YA8
 
 | | Cdc7-Dbf4 bound to ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
