5BJV
 
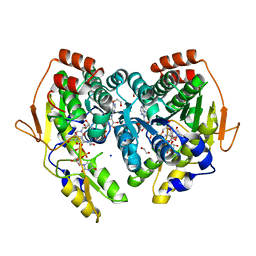 | | X-ray structure of the PglF UDP-N-acetylglucosamine 4,6-dehydratase from Campylobacterjejuni, D396N/K397A variant in complex with UDP-N-acrtylglucosamine | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Riegert, A.S, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-09-12 | | Release date: | 2017-11-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Investigation of PglF from Campylobacter jejuni Reveals a New Mechanism for a Member of the Short Chain Dehydrogenase/Reductase Superfamily.
Biochemistry, 56, 2017
|
|
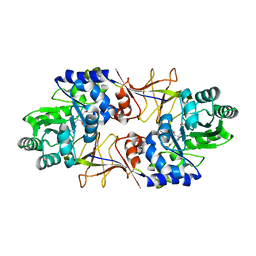
2ODO
 
 | |
3IO2
 
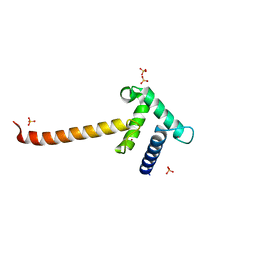 | | Crystal structure of the Taz2 domain of p300 | | Descriptor: | Histone acetyltransferase p300, SULFATE ION, ZINC ION | | Authors: | Miller, M, Dauter, Z, Wlodawer, A. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Taz2 domain of p300: insights into ligand binding.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4ZSD
 
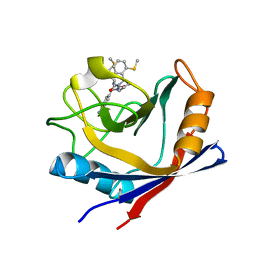 | | Human Cyclophilin D Complexed with an Inhibitor at room temperature | | Descriptor: | 1-(4-aminobenzyl)-3-[(2S)-4-(methylsulfanyl)-1-{(2R)-2-[2-(methylsulfanyl)phenyl]pyrrolidin-1-yl}-1-oxobutan-2-yl]urea, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Gelin, M, Delfosse, V, Allemand, F, Hoh, F, Sallaz-Damaz, Y, Pirocchi, M, Bourguet, W, Ferrer, J.-L, Labesse, G, Guichou, J.-F. | | Deposit date: | 2015-05-13 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Combining `dry' co-crystallization and in situ diffraction to facilitate ligand screening by X-ray crystallography.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1W9T
 
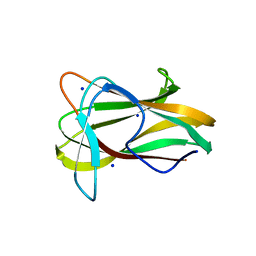 | | Structure of a beta-1,3-glucan binding CBM6 from Bacillus halodurans in complex with xylobiose | | Descriptor: | BH0236 PROTEIN, SODIUM ION, alpha-D-xylopyranose, ... | | Authors: | Boraston, A.B, van Bueren, A.L. | | Deposit date: | 2004-10-18 | | Release date: | 2004-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Family 6 Carbohydrate Binding Modules Recognize the Non-Reducing End of Beta-1,3-Linked Glucans by Presenting a Unique Ligand Binding Surface
J.Biol.Chem., 280, 2005
|
|
2JOA
 
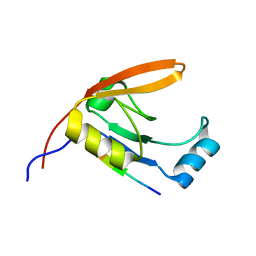 | | HtrA1 bound to an optimized peptide: NMR assignment of PDZ domain and ligand resonances | | Descriptor: | Peptide H1-C1, Serine protease HTRA1 | | Authors: | Runyon, S.T, Zhang, Y, Appleton, B.A, Sazinksy, S.L, Wu, P, Pan, B, Wiesmann, C, Skelton, N.J, Sidhu, S.S. | | Deposit date: | 2007-03-01 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the PDZ domains of human HtrA1 and HtrA3
Protein Sci., 16, 2007
|
|
8VV1
 
 | | Estrogen receptor alpha ligand binding domain in complex with palazestrant | | Descriptor: | Estrogen receptor, GLYCEROL, palazestrant | | Authors: | Ng, R.A, Barratt, S, Parisian, A, Palanisamy, G, Sun, R, Robello, B, Pena, G, Sapugay, J, Yeghikyan, D, Duncan, A, Andersen, S.E, Chawla, R, Rich, B, Hearn, B, Harmon, C, Hodges-Gallagher, L, Kushner, P.J, Greene, G.L, Myles, D.C, Fanning, S.W. | | Deposit date: | 2024-01-30 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.196 Å) | | Cite: | Estrogen receptor alpha ligand binding domain in complex with palazestrant
To Be Published
|
|
1IUY
 
 | |
5FII
 
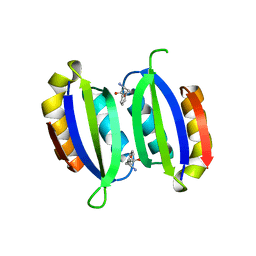 | | Structure of a human aspartate kinase, chorismate mutase and TyrA domain. | | Descriptor: | PHENYLALANINE, PHENYLALANINE-4-HYDROXYLASE | | Authors: | Patel, D, Kopec, J, Shrestha, L, Fitzpatrick, F, Pinkas, D, Chaikuad, A, Dixon-Clarke, S, McCorvie, T.J, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Ligand-Dependent Dimerization of Phenylalanine Hydroxylase Regulatory Domain.
Sci.Rep., 6, 2016
|
|
1RFG
 
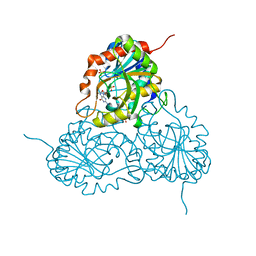 | | Crystal Structure of Human Purine Nucleoside Phosphorylase Complexed with Guanosine | | Descriptor: | GUANOSINE, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Canduri, F, Silva, R.G, Dos Santos, D.M, Palma, M.S, Basso, L.A, Santos, D.S, de Azevedo, W.F. | | Deposit date: | 2003-11-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human PNP complexed with ligands.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4E8G
 
 | | Crystal structure of an enolase (mandelate racemase subgroup) from paracococus denitrificans pd1222 (target nysgrc-012907) with bound mg | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme, N-terminal domain protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Sojitra, S, Chamala, S, Kar, A, Lafleur, J, Villigas, G, Evans, B, Hammonds, J, Gizzi, A, Zencheck, W.D, Hillerich, B, Love, J, Seidel, R.D, Bonanno, J.B, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Prediction and biochemical demonstration of a catabolic pathway for the osmoprotectant proline betaine.
MBio, 5, 2014
|
|
8X6F
 
 | | Cryo-EM structure of Staphylococcus aureus sigA-dependent RNAP-promoter open complex | | Descriptor: | DNA (71-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Yuan, L, Xu, L, Liu, Q, Feng, Y. | | Deposit date: | 2023-11-21 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of promoter recognition by Staphylococcus aureus RNA polymerase.
Nat Commun, 15, 2024
|
|
2GN2
 
 | |
5DPF
 
 | | Thermolysin in complex with inhibitor. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-09-12 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Thermodynamics of protein-ligand interactions as a reference for computational analysis: how to assess accuracy, reliability and relevance of experimental data.
J. Comput. Aided Mol. Des., 29, 2015
|
|
6W25
 
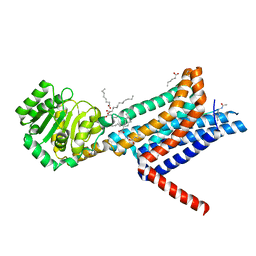 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
3QLX
 
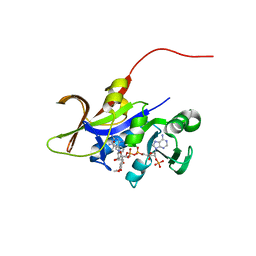 | | Candida glabrata dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.239 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
3A1Z
 
 | | Crystal structure of juvenile hormone binding protein from silkworm | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hemolymph juvenile hormone binding protein, ZINC ION | | Authors: | Suzuki, R, Fujimoto, Z, Shiotsuki, T, Momma, M, Tase, A, Yamazaki, T. | | Deposit date: | 2009-04-27 | | Release date: | 2010-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of silkworm Bombyx mori JHBP in complex with 2-methyl-2,4-pentanediol: plasticity of JH-binding pocket and ligand-induced conformational change of the second cavity in JHBP
Plos One, 8, 2013
|
|
1K2F
 
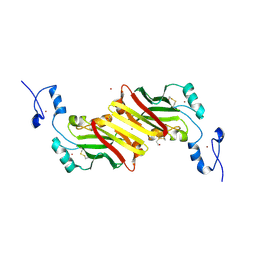 | | siah, Seven In Absentia Homolog | | Descriptor: | BETA-MERCAPTOETHANOL, ZINC ION, siah-1A protein | | Authors: | Polekhina, G, House, C.M, Traficante, N, Mackay, J.P, Relaix, F, Sassoon, D.A, Parker, M.W, Bowtell, D.D.L. | | Deposit date: | 2001-09-26 | | Release date: | 2001-12-28 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling.
Nat.Struct.Biol., 9, 2002
|
|
5KX9
 
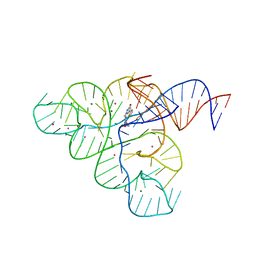 | | Selective Small Molecule Inhibition of the FMN Riboswitch | | Descriptor: | (6P)-2-{(3S)-1-[(2-methoxypyrimidin-5-yl)methyl]piperidin-3-yl}-6-(thiophen-2-yl)pyrimidin-4-ol, FMN Riboswitch, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-07-20 | | Release date: | 2016-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atomic resolution mechanistic studies of ribocil: A highly selective unnatural ligand mimic of the E. coli FMN riboswitch.
Rna Biol., 13, 2016
|
|
3FWG
 
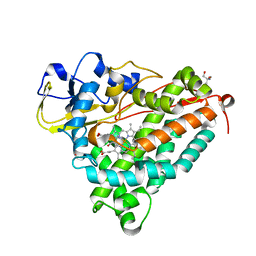 | | Ferric camphor bound Cytochrome P450cam, Arg365Leu, Glu366Gln, monoclinic crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, Camphor 5-monooxygenase, ... | | Authors: | Schlichting, I, Von Koenig, K, Aldag, C, Hilvert, D. | | Deposit date: | 2009-01-18 | | Release date: | 2009-03-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1P2H
 
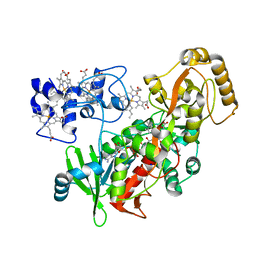 | | H61M mutant of flavocytochrome c3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALATE LIKE INTERMEDIATE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Rothery, E.L, Mowat, C.G, Miles, C.S, Walkinshaw, M.D, Reid, G.A, Chapman, S.K. | | Deposit date: | 2003-04-15 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Histidine 61: An Important Heme Ligand in the Soluble Fumarate Reductase from Shewanella
frigidimarina
Biochemistry, 42, 2003
|
|
7U3G
 
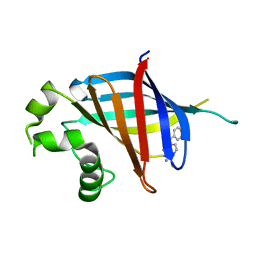 | | GID4 in complex with compound 67 | | Descriptor: | (1R)-1-phenyl-1,2,3,4-tetrahydroisoquinolin-5-amine, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
7U3K
 
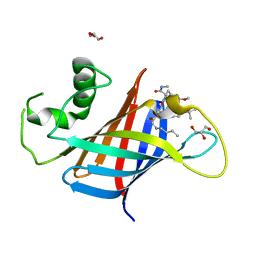 | | GID4 in complex with compound 89 | | Descriptor: | GLYCEROL, Glucose-induced degradation protein 4 homolog, N-butylglycyl-4-tert-butyl-D-phenylalanyl-3-methoxy-N-methyl-L-phenylalaninamide | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
3QLS
 
 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP115A) | | Descriptor: | 6-methyl-5-[3-methyl-3-(3,4,5-trimethoxyphenyl)but-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
7U3I
 
 | | GID4 in complex with compound 16 | | Descriptor: | 3-{[(5S)-5-methyl-4,5-dihydro-1,3-thiazol-2-yl]amino}phenol, GLYCEROL, Glucose-induced degradation protein 4 homolog | | Authors: | Chana, C.K, Sicheri, F. | | Deposit date: | 2022-02-27 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.991 Å) | | Cite: | Discovery and Structural Characterization of Small Molecule Binders of the Human CTLH E3 Ligase Subunit GID4.
J.Med.Chem., 65, 2022
|
|
