1MXT
 
 | | Atomic resolution structure of Cholesterol oxidase (Streptomyces sp. SA-COO) | | Descriptor: | CHOLESTEROL OXIDASE, FLAVIN-N7 PROTONATED-ADENINE DINUCLEOTIDE, OXYGEN MOLECULE, ... | | Authors: | Vrielink, A, Lario, P.I. | | Deposit date: | 2002-10-03 | | Release date: | 2003-02-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Sub-atomic resolution crystal structure of cholesterol oxidase:
What atomic resolution crystallography reveals about enzyme mechanism and the role of FAD cofactor in redox activity
J.Mol.Biol., 326, 2003
|
|
8SMV
 
 | | GPR161 Gs heterotrimer | | Descriptor: | CHOLESTEROL, G-protein coupled receptor 161, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Hoppe, N, Manglik, A, Harrison, S. | | Deposit date: | 2023-04-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | GPR161 structure uncovers the redundant role of sterol-regulated ciliary cAMP signaling in the Hedgehog pathway.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1RTL
 
 | | CRYSTAL STRUCTURE OF HCV NS3 PROTEASE DOMAIN: NS4A PEPTIDE COMPLEX WITH COVALENTLY BOUND PYRROLIDINE-5,5-TRANSLACTAM INHIBITOR | | Descriptor: | N-[(2R,3S)-1-((2S)-2-{[(CYCLOPENTYLAMINO)CARBONYL]AMINO}-3-METHYLBUTANOYL)-2-(1-FORMYL-1-CYCLOBUTYL)PYRROLIDINYL]CYCLOPROPANECARBOXAMIDE, NS3 protease/helicase, NS4A COFACTOR, ... | | Authors: | Skarzynski, T, Somers, D.O.N. | | Deposit date: | 2003-12-10 | | Release date: | 2004-12-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Pyrrolidine-5,5-trans-lactams. 4. Incorporation of a P3/P4 urea leads to potent intracellular inhibitors of hepatitis C virus NS3/4A protease
Org.Lett., 5, 2003
|
|
1RPX
 
 | |
1RXZ
 
 | | C-terminal region of A. fulgidus FEN-1 complexed with A. fulgidus PCNA | | Descriptor: | DNA polymerase sliding clamp, Flap structure-specific endonuclease | | Authors: | Chapados, B.R, Hosfield, D.J, Han, S, Qiu, J, Yelent, B, Shen, B, Tainer, J.A. | | Deposit date: | 2003-12-18 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for FEN-1 Substrate Specificity and PCNA-Mediated Activation in DNA Replication and Repair
Cell(Cambridge,Mass.), 116, 2004
|
|
3G4P
 
 | | OXA-24 beta-lactamase at pH 7.5 | | Descriptor: | Beta-lactamase OXA-24 | | Authors: | Santillana, E, Romero, A. | | Deposit date: | 2009-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, synthesis, and crystal structures of 6-alkylidene-2'-substituted penicillanic acid sulfones as potent inhibitors of Acinetobacter baumannii OXA-24 carbapenemase
J.Am.Chem.Soc., 132, 2010
|
|
3GGV
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-3-hydroxy-4-{[(2S)-2-(3-{[6-(1-hydroxy-1-methylethyl)pyridin-2-yl]methyl}-2-oxo-2,3-dihydro-1H-imidazol-1-yl)-3,3-dimethylbutanoyl]amino}-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
3G86
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with thiazine inhibitor | | Descriptor: | N-{3-[6-fluoro-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2-dihydroquinolin-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, NICKEL (II) ION, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-02-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 2: Synthesis and structure-activity relationships of benzothiazine-substituted quinolinediones
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1LY4
 
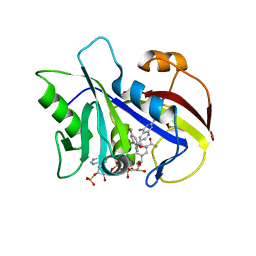 | | Analysis of quinazoline and PYRIDO[2,3D]PYRIMIDINE N9-C10 reversed bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase | | Descriptor: | 2,4-DIAMINO-6-[N-(3',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]PYRIDO[2,3-D]PYRIMIDINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1LY3
 
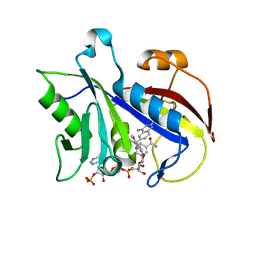 | | ANALYSIS OF QUINAZOLINE AND PYRIDOPYRIMIDINE N9-C10 REVERSED BRIDGE ANTIFOLATES IN COMPLEX WITH NADP+ AND PNEUMOCYSTIS CARINII DIHYDROFOLATE REDUCTASE | | Descriptor: | 2,4-DIAMINO-6-[N-(2',5'-DIMETHOXYBENZYL)-N-METHYLAMINO]QUINAZOLINE, DIHYDROFOLATE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Galitsky, N, Luft, J.R, Pangborn, W, Queener, S.F, Gangjee, A. | | Deposit date: | 2002-06-06 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of quinazoline and pyrido[2,3-d]pyrimidine N9-C10 reversed-bridge antifolates in complex with NADP+ and Pneumocystis carinii dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
3GGA
 
 | | HIV Protease inhibitors with pseudo-symmetric cores | | Descriptor: | V-1 protease, methyl [(1S,4S,5S,7S,10S)-4-benzyl-1,10-di-tert-butyl-5-hydroxy-2,9,12-trioxo-7-(4-pyridin-2-ylbenzyl)-13-oxa-3,8,11-triazatetradec-1-yl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
6IMO
 
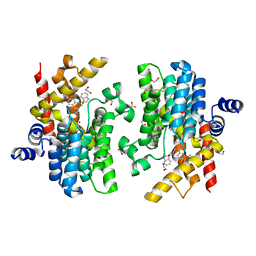 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[(1H-indol-3-yl)methyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3I3S
 
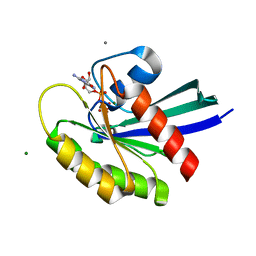 | | Crystal Structure of H-Ras with Thr50 replaced by Isoleucine | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Gremer, L, Dvorsky, R, Merbitz-Zahradnik, T, Wittinghofer, A, Ahmadian, M.R. | | Deposit date: | 2009-06-30 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A restricted spectrum of NRAS mutations causes Noonan syndrome.
Nat.Genet., 42, 2010
|
|
1O5U
 
 | |
3GWA
 
 | |
3GNV
 
 | | HCV NS5B polymerase in complex with 1,5 benzodiazepine inhibitor 1b | | Descriptor: | (11R)-10-acetyl-11-[4-(benzyloxy)-2-chlorophenyl]-6-hydroxy-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, GLYCEROL, RNA-directed RNA polymerase | | Authors: | De Bondt, H, Nyanguile, O. | | Deposit date: | 2009-03-18 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of a benzodiazepine scaffold yields a potent allosteric inhibitor of hepatitis C NS5B RNA polymerase.
J.Med.Chem., 52, 2009
|
|
3GWE
 
 | |
3H5U
 
 | | Hepatitis C virus polymerase NS5B with saccharin inhibitor 1 | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzisothiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 4: structure-based design, synthesis, and biological evaluation of benzo[d]isothiazole-1,1-dioxides
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3H59
 
 | | Hepatitis C virus polymerase NS5B with thiazine inhibitor 2 | | Descriptor: | N-{3-[(5S)-5-(1,1-dimethylpropyl)-1-(4-fluoro-3-methylbenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-4H-1,4-benzothiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Ghate, M. | | Deposit date: | 2009-04-21 | | Release date: | 2009-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of HCV polymerase NS5B. Part 3: synthesis and optimization studies of benzothiazine-substituted tetramic acids
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HVG
 
 | | Structure of bace (beta secretase) in Complex with EV0 | | Descriptor: | 2-amino-6-propylpyrimidin-4(3H)-one, Beta-secretase 1, GLYCEROL | | Authors: | Godemann, R, Madden, J, Kramer, J, Smith, M.A, Barker, J, Ebneth, A. | | Deposit date: | 2009-06-16 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Fragment-Based Discovery of BACE1 Inhibitors Using Functional Assays
Biochemistry, 48, 2009
|
|
3HLN
 
 | | Crystal structure of ClpP A153C mutant with inter-heptamer disulfide bonds | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, CALCIUM ION | | Authors: | Kimber, M.S, Yu, A.Y.H, Borg, M, Chan, H.S, Houry, W.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Theoretical Studies Indicate that the Cylindrical Protease ClpP Samples Extended and Compact Conformations.
Structure, 18, 2010
|
|
3HW1
 
 | | Structure of Bace (beta secretase) in complex with ligand EV2 | | Descriptor: | 3-pyrrolidin-1-ylquinoxalin-2-amine, Beta-secretase 1, GLYCEROL | | Authors: | Godemann, R, Madden, J, Kramer, J, Smith, M.A, Barker, J, Ebneth, A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Fragment-Based Discovery of BACE1 Inhibitors Using Functional Assays
Biochemistry, 48, 2009
|
|
6INM
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[2-(1H-indol-3-yl)ethyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
6IMR
 
 | | Crystal structure of PDE4D complexed with a novel inhibitor | | Descriptor: | (1S)-1-[3-(1H-indol-3-yl)propyl]-6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-carbaldehyde, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Zhang, X.L, Su, H.X, Xu, Y.C. | | Deposit date: | 2018-10-23 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structure-Aided Identification and Optimization of Tetrahydro-isoquinolines as Novel PDE4 Inhibitors Leading to Discovery of an Effective Antipsoriasis Agent.
J.Med.Chem., 62, 2019
|
|
3EZO
 
 | |
