6FOB
 
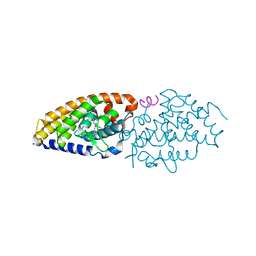 | | Vitamin D receptor complex 5 | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-heptyl)phenyl]dec-2-enylidene]cyclohexane-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2018-02-06 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aromatic-Based Design of Highly Active and Noncalcemic Vitamin D Receptor Agonists.
J. Med. Chem., 61, 2018
|
|
8QK2
 
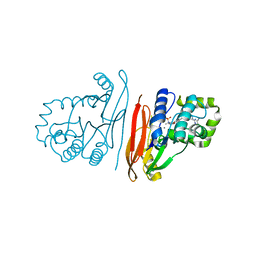 | | Structure of K.pneumoniae LpxH in complex with EBL-3339 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK9
 
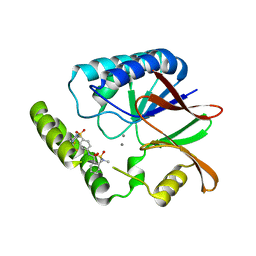 | | Structure of E. coli LpxH in complex with JEDI-1444 | | Descriptor: | 2-[methyl(methylsulfonyl)amino]-~{N}-[4-[4-[3-(trifluoromethyl)phenyl]piperazin-1-yl]sulfonylphenyl]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8QK5
 
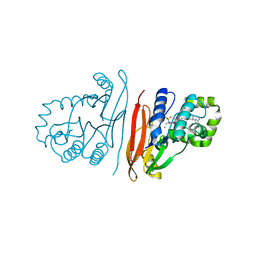 | | Structure of K. pneumoniae LpxH in complex with EBL-3647 | | Descriptor: | 5-[[3-(aminomethyl)azetidin-1-yl]methyl]-N-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6GK0
 
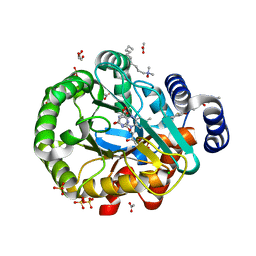 | | HUMAN DIHYDROOROTATE DEHYDROGENASE IN COMPLEX WITH CLASS III HISTONE DEACETYLASE INHIBITOR | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 4-~{tert}-butyl-~{N}-[[4-[5-(dimethylamino)pentanoylamino]phenyl]carbamothioyl]benzamide, ACETIC ACID, ... | | Authors: | Hakansson, M, Ladds, M.J.G.W, Walse, B, Lain, S. | | Deposit date: | 2018-05-17 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploitation of dihydroorotate dehydrogenase (DHODH) and p53 activation as therapeutic targets: A case study in polypharmacology.
J.Biol.Chem., 295, 2020
|
|
3MKP
 
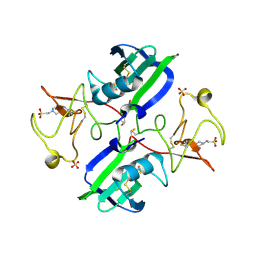 | | Crystal structure of 1K1 mutant of Hepatocyte Growth Factor/Scatter Factor fragment NK1 in complex with heparin | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gherardi, E, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2010-04-15 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Engineering a fragment of Hepatocyte Growth Factor/Scatter Factor for tissue and organ regeneration
To be Published
|
|
6UUO
 
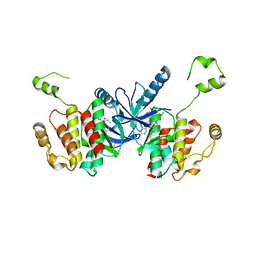 | | Crystal structure of BRAF kinase domain bound to the PROTAC P4B | | Descriptor: | N-(3-{5-[(1-acetylpiperidin-4-yl)(methyl)amino]-3-(pyrimidin-5-yl)-1H-pyrrolo[3,2-b]pyridin-1-yl}-2,4-difluorophenyl)propane-1-sulfonamide, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Posternak, G, Kurinov, I, Sicheri, F. | | Deposit date: | 2019-10-30 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Functional characterization of a PROTAC directed against BRAF mutant V600E.
Nat.Chem.Biol., 16, 2020
|
|
7KDT
 
 | |
4LM8
 
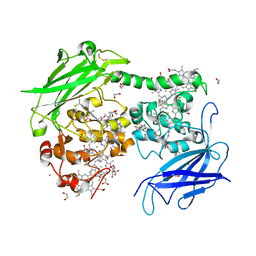 | |
7LXT
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound bortezomib | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Morton, C.J, Metcalfe, R.D, Liu, B, Xie, S.C, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LXU
 
 | | Structure of Plasmodium falciparum 20S proteasome with bound MPI-5 | | Descriptor: | 20S proteasome alpha-1 subunit, 20S proteasome alpha-2 subunit, 20S proteasome alpha-3 subunit, ... | | Authors: | Metcalfe, R.D, Morton, C.J, Xie, S.C, Liu, B, Hanssen, E, Leis, A.P, Tilley, L, Griffin, M.D.W. | | Deposit date: | 2021-03-05 | | Release date: | 2021-09-22 | | Last modified: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Design of proteasome inhibitors with oral efficacy in vivo against Plasmodium falciparum and selectivity over the human proteasome.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LGV
 
 | |
1RCQ
 
 | | The 1.45 A crystal structure of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms | | Descriptor: | D-LYSINE, PYRIDOXAL-5'-PHOSPHATE, catabolic alanine racemase DadX | | Authors: | Le Magueres, P, Im, H, Dvorak, A, Strych, U, Benedik, M, Krause, K.L. | | Deposit date: | 2003-11-04 | | Release date: | 2004-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45 A resolution of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms.
Biochemistry, 42, 2003
|
|
3QIX
 
 | | Crystal Structure of BoNT/A LC with Zinc bound | | Descriptor: | 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ZINC ION | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.413 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QJ0
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-3 | | Descriptor: | (4R)-4-(4-chlorophenoxy)-1-[(4-chlorophenyl)sulfonyl]-N-hydroxy-L-prolinamide, 1,2-ETHANEDIOL, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QIZ
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-2 | | Descriptor: | (2S,4R)-2-(2-{[3-(4-fluoro-3-methylphenyl)propyl](methyl)amino}ethyl)-4-(4-fluorophenyl)-N-hydroxy-4-methoxybutanamide, Botulinum neurotoxin type A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
3QLQ
 
 | | Crystal structure of Concanavalin A bound to an octa-alpha-mannosyl-octasilsesquioxane cluster | | Descriptor: | CALCIUM ION, Concanavalin-A, MANGANESE (II) ION, ... | | Authors: | Sundberg, E.J, Bonsor, D.A, Trastoy, B, Chiara, J.L. | | Deposit date: | 2011-02-03 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and Biophysical Study of Disassembling Nano hybrid Bioconjugates with a Cubic Octasilsesquioxane Core
Adv Funct Mater, 22, 2012
|
|
3QIY
 
 | | Crystal Structure of BoNT/A LC complexed with Hydroxamate-based Inhibitor PT-1 | | Descriptor: | 1,2-ETHANEDIOL, 4-[bis(4-chlorobenzyl)amino]-N-hydroxybutanamide, Botulinum neurotoxin type A, ... | | Authors: | Thompson, A.A, Han, G.W, Stevens, R.C. | | Deposit date: | 2011-01-28 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Three Novel Hydroxamate-Based Zinc Chelating Inhibitors of the Clostridium botulinum Serotype A Neurotoxin Light Chain Metalloprotease Reveals a Compact Binding Site Resulting from 60/70 Loop Flexibility.
Biochemistry, 50, 2011
|
|
4BY4
 
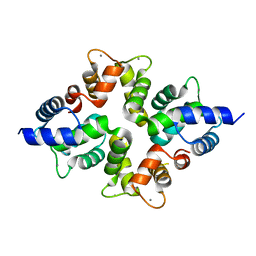 | |
4BY5
 
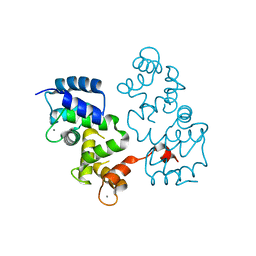 | |
4P18
 
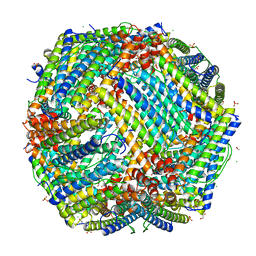 | | Crystal Structure of frog M ferritin mutant D80K | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Ghini, V, Turano, P. | | Deposit date: | 2014-02-25 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Loop electrostatics modulates the intersubunit interactions in ferritin.
Acs Chem.Biol., 9, 2014
|
|
3VVA
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with ascofuranone derivative | | Descriptor: | 3-chloro-4,6-dihydroxy-5-[(2E,6E,8S)-8-hydroxy-3,7-dimethylnona-2,6-dien-1-yl]-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Kido, Y, Sakamoto, K, Inaoka, D.K, Tsuge, C, Tatsumi, R, Balogun, E.O, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Saimoto, H, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2012-07-17 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of the trypanosome cyanide-insensitive alternative oxidase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HPG
 
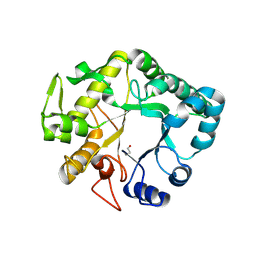 | | Crystal structure of a glycosylated beta-1,3-glucanase (HEV B 2), an allergen from Hevea brasiliensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rodriguez-Romero, A, Hernandez-Santoyo, A. | | Deposit date: | 2012-10-23 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5364 Å) | | Cite: | Structural analysis of the endogenous glycoallergen Hev b 2 (endo-beta-1,3-glucanase) from Hevea brasiliensis and its recognition by human basophils.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3W54
 
 | | Crystal structure of cyanide-insensitive alternative oxidase from Trypanosoma brucei with colletochlorin B | | Descriptor: | 3-chloro-5-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-4,6-dihydroxy-2-methylbenzaldehyde, Alternative oxidase, mitochondrial, ... | | Authors: | Shiba, T, Kido, Y, Sakamoto, K, Inaoka, D.K, Tsuge, C, Tatsumi, R, Takahashi, G, Balogun, E.O, Nara, T, Aoki, T, Honma, T, Tanaka, A, Inoue, M, Matsuoka, S, Saimoto, H, Moore, A.L, Harada, S, Kita, K. | | Deposit date: | 2013-01-21 | | Release date: | 2013-03-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the trypanosome cyanide-insensitive alternative oxidase
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IIS
 
 | | Crystal structure of a glycosylated beta-1,3-glucanase (HEV B 2), An allergen from Hevea Brasiliensis (Space group P41) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-1,3-glucanase form 'RRII Gln 2', CACODYLATE ION, ... | | Authors: | Rodriguez-Romero, A, Hernandez-Santoyo, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6676 Å) | | Cite: | Structural analysis of the endogenous glycoallergen Hev b 2 (endo-beta-1,3-glucanase) from Hevea brasiliensis and its recognition by human basophils.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
