7OVD
 
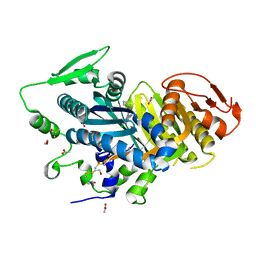 | | Human soluble adenylyl cyclase in complex with the inhibitor TDI10229 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloranyl-6-[1,5-dimethyl-4-(phenylmethyl)pyrazol-3-yl]pyrimidin-2-amine, ACETATE ION, ... | | Authors: | Steegborn, C, Quast, J. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of TDI-10229: A Potent and Orally Bioavailable Inhibitor of Soluble Adenylyl Cyclase (sAC, ADCY10).
Acs Med.Chem.Lett., 12, 2021
|
|
8JWJ
 
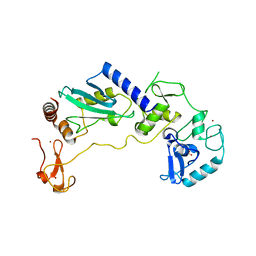 | | PHD Finger Protein 7 (PHF7) in complex with UBE2D2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHD finger protein 7, ... | | Authors: | Lee, H.S, Bang, I, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8JWS
 
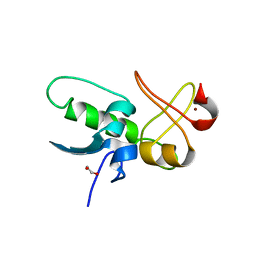 | | ePHD domain of PHD Finger Protein 7 (PHF7) | | Descriptor: | 1,2-ETHANEDIOL, PHD finger protein 7, ZINC ION | | Authors: | Bang, I, Lee, H.S, Choi, H.-J. | | Deposit date: | 2023-06-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for PHF7-mediated ubiquitination of histone H3.
Genes Dev., 37, 2023
|
|
8JWU
 
 | |
8OQ5
 
 | |
8OQ4
 
 | |
8K7Y
 
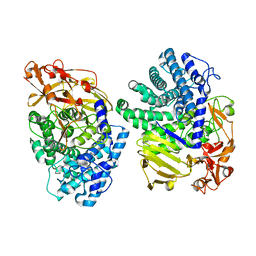 | | Crystal structure of GH146 beta-L-arabinofuranosidase Bll3HypBA1 (amino acids 380-1051), ligand-free form | | Descriptor: | ZINC ION, beta1,3-L-arabinofuranoside | | Authors: | Maruyama, S, Pan, L, Miyake, M, Fujita, K, Fushinobu, S. | | Deposit date: | 2023-07-27 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Bifidobacterial GH146 beta-L-arabinofuranosidase for the removal of beta 1,3-L-arabinofuranosides on plant glycans.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
7PCW
 
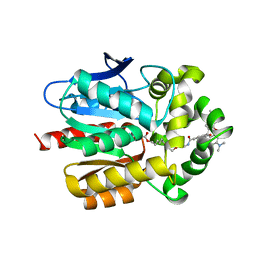 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-M175W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-(6-chloranylhexoxy)ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7PCX
 
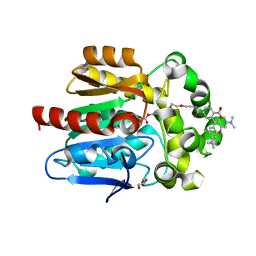 | | X-RAY STRUCTURE OF THE HALOALKANE DEHALOGENASE HALOTAG7-Q165W LABELED WITH A CHLOROALKANE-TETRAMETHYLRHODAMINE FLUOROPHORE SUBSTRATE | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Frei, M, Hiblot, J, Johnsson, K. | | Deposit date: | 2021-08-04 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Engineered HaloTag variants for fluorescence lifetime multiplexing.
Nat.Methods, 19, 2022
|
|
7OEC
 
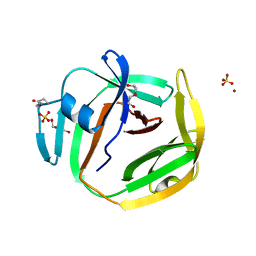 | | Crystal structure of an intein from a hyperthermophile | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA polymerase II large subunit, SULFATE ION, ... | | Authors: | Hannes, B, Hiltunen, M, Iwai, H. | | Deposit date: | 2021-05-03 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mini-Intein Structures from Extremophiles Suggest a Strategy for Finding Novel Robust Inteins.
Microorganisms, 9, 2021
|
|
8JVI
 
 | | Structure of human TRPV4 with antagonist A2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JU5
 
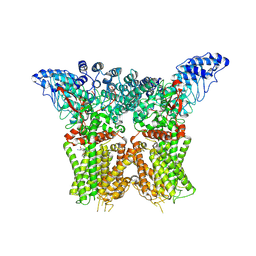 | | Structure of human TRPV4 with antagonist A1 | | Descriptor: | 4-[(3~{S},4~{S})-4-(aminomethyl)-1-(5-chloranylpyridin-2-yl)sulfonyl-4-oxidanyl-pyrrolidin-3-yl]oxy-2-fluoranyl-benzenecarbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JU6
 
 | | Structure of human TRPV4 with antagonist GSK279 | | Descriptor: | 1-({(5S,7S)-3-[5-(2-hydroxypropan-2-yl)pyrazin-2-yl]-7-methyl-2-oxo-1-oxa-3-azaspiro[4.5]decan-7-yl}methyl)-1H-benzimidazole-6-carbonitrile, Transient receptor potential cation channel subfamily V member 4,3C-GFP | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-24 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
8JVJ
 
 | | Structure of human TRPV4 with antagonist A2 and RhoA | | Descriptor: | Transforming protein RhoA, Transient receptor potential cation channel subfamily V member 4,3C-GFP, [6-[[4-(2,4-dimethyl-1,3-thiazol-5-yl)-1,3-thiazol-2-yl]amino]pyridin-3-yl]-[(1~{S},5~{R})-3-[5-(trifluoromethyl)pyrimidin-2-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Fan, J, Lei, X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural Pharmacology of TRPV4 Antagonists.
Adv Sci, 11, 2024
|
|
7FYA
 
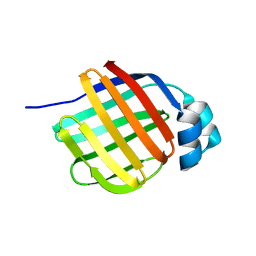 | |
7G1X
 
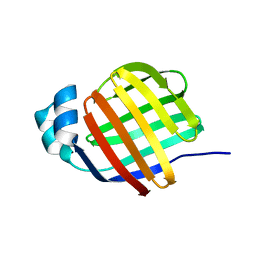 | |
7FXO
 
 | | Crystal Structure of apo human FABP1 monoclinic form III | | Descriptor: | BENZAMIDINE, DIMETHYL SULFOXIDE, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of a human FABP1 complex
To be published
|
|
7O0K
 
 | |
7O0J
 
 | |
7G0W
 
 | | Crystal Structure of apo human FABP1i monoclinic form II, twinned with beta=90deg | | Descriptor: | BENZAMIDINE, CHLORIDE ION, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of a human FABP1 complex
To be published
|
|
7FY8
 
 | | Crystal Structure of human FABP1 in complex with 2-[[4-cyclopropyl-5-methyl-3-[3-(trifluoromethyl)-1,2,4-oxadiazol-5-yl]thiophen-2-yl]carbamoyl]cyclopentene-1-carboxylic acid | | Descriptor: | 2-({(3P)-4-cyclopropyl-5-methyl-3-[3-(trifluoromethyl)-1,2,4-oxadiazol-5-yl]thiophen-2-yl}carbamoyl)cyclopent-1-ene-1-carboxylic acid, Fatty acid-binding protein, liver, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Richter, H, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal Structure of a human FABP1 complex
To be published
|
|
8OV0
 
 | | MUC5AC CysD7 amino acids 3518-3626 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mucin-5AC | | Authors: | Khmelnitsky, L, Milo, A, Dym, O, Fass, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity of CysD domains in gel-forming mucins.
Febs J., 290, 2023
|
|
7P2Z
 
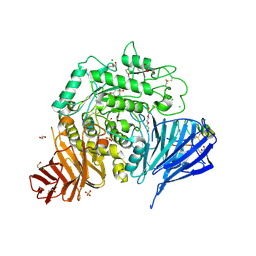 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 4 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7P32
 
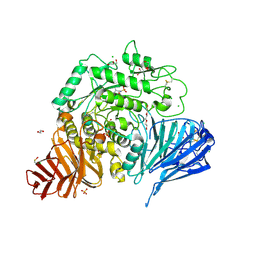 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
7E32
 
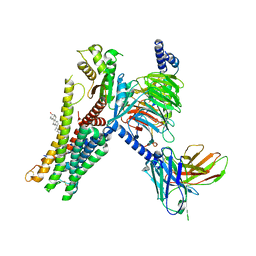 | | Serotonin 1D (5-HT1D) receptor-Gi protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, P, Huang, S, Zhang, H, Mao, C, Zhou, X.E, Shen, D.D, Jiang, Y, Zhang, Y, Xu, H.E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-04-21 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into the lipid and ligand regulation of serotonin receptors.
Nature, 592, 2021
|
|
