8W8T
 
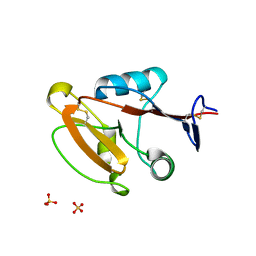 | | Crystal structure of human CLEC12A CRD | | Descriptor: | C-type lectin domain family 12 member A, SULFATE ION | | Authors: | Mori, S, Nagae, M, Yamasaki, S. | | Deposit date: | 2023-09-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of CLEC12A and an antibody that interferes with binding of diverse ligands.
Int.Immunol., 36, 2024
|
|
3M0Q
 
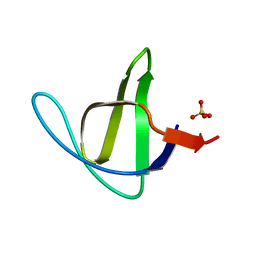 | |
4Y5A
 
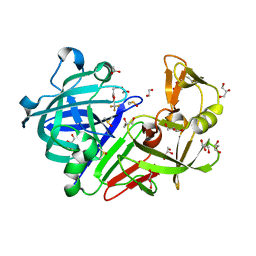 | | Endothiapepsin in complex with fragment 206 | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-(trifluoromethyl)furan-2-yl]methanamine, ACETATE ION, ... | | Authors: | Ehrmann, F.R, Huschmann, F.U, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Fragment Sreening of an Entire Library
To Be Published
|
|
6QEN
 
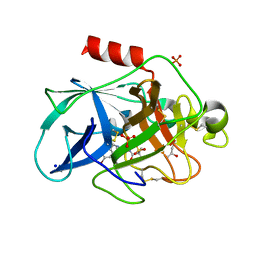 | | Crystal structure of Porcine Pancreatic Elastase (PPE) in complex with the 3-Oxo-beta-Sultam inhibitor LMC240 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-[[1-[(4-bromophenyl)methyl]-1,2,3-triazol-4-yl]methylamino]-2-methyl-1-oxidanylidene-propane-2-sulfonic acid, Chymotrypsin-like elastase family member 1, ... | | Authors: | Brito, J.A, Almeida, V.T, Carvalho, L.M, Moreira, R, Archer, M. | | Deposit date: | 2019-01-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.195 Å) | | Cite: | 3-Oxo-beta-sultam as a Sulfonylating Chemotype for Inhibition of Serine Hydrolases and Activity-Based Protein Profiling.
Acs Chem.Biol., 15, 2020
|
|
4Y5L
 
 | | Endothiapepsin in its apo form | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Endothiapepsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6QE1
 
 | | P38 alpha complex with AR117046 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{R})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | P38 alpha complex with AR117045 and AR117046
To Be Published
|
|
6QFE
 
 | | Crystal Structure of Human Kallikrein 5 in complex with GSK144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(5-phenyl-1~{H}-imidazol-2-yl)methylamino]-2-(pyridin-3-ylmethoxy)benzenecarboximidamide, ... | | Authors: | Thorpe, J.H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Evaluation of a crystallographic surrogate for kallikrein 5 in the discovery of novel inhibitors for Netherton syndrome.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3MDF
 
 | | Crystal structure of the RRM domain of Cyclophilin 33 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase E | | Authors: | Hom, R.A, Chang, P.Y, Roy, S, Mussleman, C.A, Glass, K.C, Seleznevia, A.I, Gozani, O, Ismagilov, R.F, Cleary, M.L, Kutateladze, T.G. | | Deposit date: | 2010-03-30 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular mechanism of MLL PHD3 and RNA recognition by the Cyp33 RRM domain.
J.Mol.Biol., 400, 2010
|
|
6QMR
 
 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
3MDM
 
 | | Thioperamide complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, N-cyclohexyl-4-(1H-imidazol-5-yl)piperidine-1-carbothioamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
6QQ3
 
 | | The room temperature structure of lysozyme via the acoustic levitation of a droplet | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Axford, D.N, Docker, P, Dye, E, Morris, R. | | Deposit date: | 2019-02-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Non-Contact Universal Sample Presentation for Room Temperature Macromolecular Crystallography Using Acoustic Levitation.
Sci Rep, 9, 2019
|
|
6QQE
 
 | |
4Y83
 
 | | Crystal structure of COT kinase domain in complex with 5-(2-amino-5-(quinolin-3-yl)pyridin-3-yl)-1,3,4-oxadiazole-2(3H)-thione | | Descriptor: | 5-[2-amino-5-(quinolin-3-yl)pyridin-3-yl]-1,3,4-oxadiazole-2(3H)-thione, Mitogen-activated protein kinase kinase kinase 8 | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2015-02-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The Crystal Structure of Cancer Osaka Thyroid Kinase Reveals an Unexpected Kinase Domain Fold.
J.Biol.Chem., 290, 2015
|
|
3MHW
 
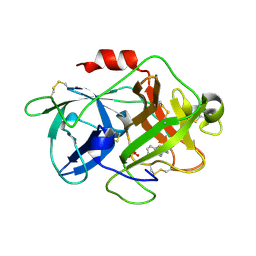 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
6QPE
 
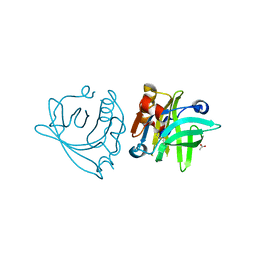 | |
4YCE
 
 | |
8V2Z
 
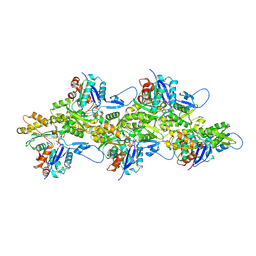 | | Cryo-EM Structure of Smooth Muscle Gamma Actin (ACTG2) Mutant R257C | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, gamma-enteric smooth muscle, ... | | Authors: | Palmer, N.J, Carman, P.J, Ceron, R.H, Dominguez, R. | | Deposit date: | 2023-11-25 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Smooth Muscle Gamma Actin (ACTG2) Filament Mutant R257C
To Be Published
|
|
4YCT
 
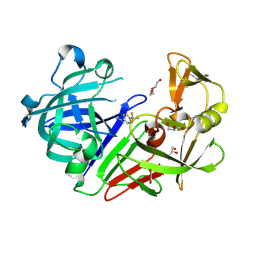 | |
4YD3
 
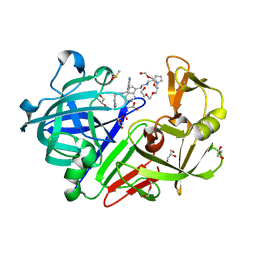 | | Endothiapepsin in complex with fragment 224 | | Descriptor: | 2-(1H-indol-3-yl)-N-[(1-methyl-1H-pyrrol-2-yl)methyl]ethanamine, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stieler, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-20 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.248 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
6QRH
 
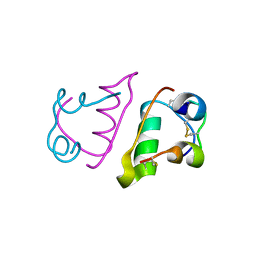 | |
8VM2
 
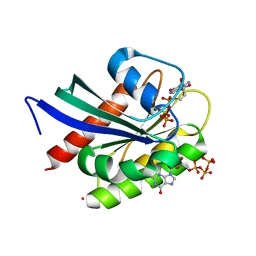 | | Crystal Structure of NRAS Q61K bound to GTP | | Descriptor: | GTPase NRas, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gebregiworgis, T, Chan, J.Y.L, Kuntz, D.A, Prive, G.G, Marshall, C.B, Ikura, M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of NRAS Q61K with a ligand-induced pocket near switch II.
Eur J Cell Biol, 103, 2024
|
|
6QRK
 
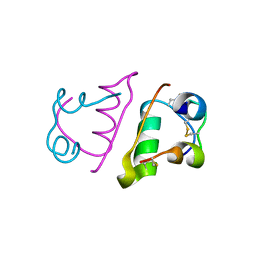 | |
3MN6
 
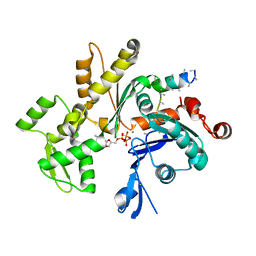 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | Authors: | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4YFR
 
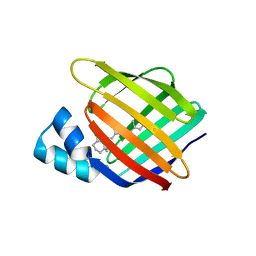 | |
6QUV
 
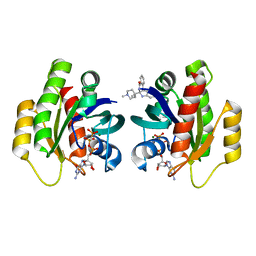 | | Crystal Structure of KRAS-G12D in complex with GMP-PCP and compound 15R | | Descriptor: | (6~{a}~{R},11~{b}~{S})-6~{a}-(1,4-dimethylpiperidin-4-yl)-7,11~{b}-dihydro-6~{H}-indolo[2,3-c]isoquinolin-5-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Fischer, G, Kessler, D, Muellauer, B, Wolkerstorfer, B. | | Deposit date: | 2019-02-28 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | KRAS Binders Hidden in Nature.
Chemistry, 25, 2019
|
|
