2XS2
 
 | |
4EWC
 
 | |
1QC0
 
 | | CRYSTAL STRUCTURE OF A 19 BASE PAIR COPY CONTROL RELATED RNA DUPLEX | | Descriptor: | 5'-R(*GP*CP*AP*CP*CP*GP*CP*UP*AP*CP*CP*AP*AP*CP*GP*GP*UP*GP*C)-3', 5'-R(*GP*CP*AP*CP*CP*GP*UP*UP*GP*GP*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', 5'-R(*UP*AP*GP*CP*GP*GP*UP*GP*C)-3', ... | | Authors: | Klosterman, P.S, Shah, S.A, Steitz, T.A. | | Deposit date: | 1999-05-17 | | Release date: | 1999-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of two plasmid copy control related RNA duplexes: An 18 base pair duplex at 1.20 A resolution and a 19 base pair duplex at 1.55 A resolution.
Biochemistry, 38, 1999
|
|
1QCU
 
 | |
3CJ5
 
 | |
1NB7
 
 | | HC-J4 RNA polymerase complexed with short RNA template strand | | Descriptor: | 5'-R(*UP*UP*UP*U)-3', MANGANESE (II) ION, polyprotein | | Authors: | O'Farrell, D.J, Trowbridge, R, Rowlands, D.J, Jaeger, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation.
J.Mol.Biol., 326, 2003
|
|
1DIX
 
 | | CRYSTAL STRUCTURE OF RNASE LE | | Descriptor: | EXTRACELLULAR RIBONUCLEASE LE | | Authors: | Tanaka, N, Nakamura, K.T. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a plant ribonuclease, RNase LE.
J.Mol.Biol., 298, 2000
|
|
5E9W
 
 | |
1EFZ
 
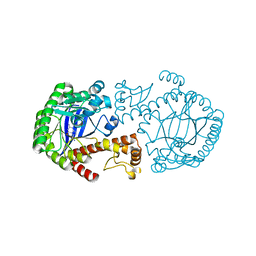 | | MUTAGENESIS AND CRYSTALLOGRAPHIC STUDIES OF ZYMOMONAS MOBILIS TRNA-GUANINE TRANSGLYCOSYLASE TO ELUCIDATE THE ROLE OF SERINE 103 FOR ENZYMATIC ACTIVITY | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Gradler, U, Ficner, R, Garcia, G.A, Stubbs, M.T, Klebe, G, Reuter, K. | | Deposit date: | 2000-02-11 | | Release date: | 2000-03-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase to elucidate the role of serine 103 for enzymatic activity.
FEBS Lett., 454, 1999
|
|
6ZWX
 
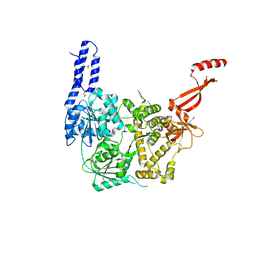 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4Q9Q
 
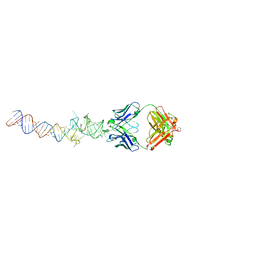 | | Crystal structure of an RNA aptamer bound to bromo-ligand analog in complex with Fab | | Descriptor: | (5Z)-5-(3-bromobenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, Fab BL3-6, HEAVY CHAIN, ... | | Authors: | Huang, H, Suslov, N.B, Li, N.-S, Shelke, S.A, Evans, M.E, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-18 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
1OS5
 
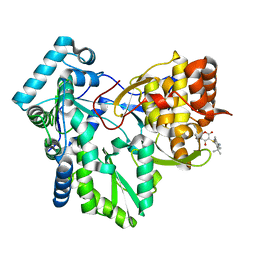 | | Crystal structure of HCV NS5B RNA polymerase complexed with a novel non-competitive inhibitor. | | Descriptor: | 3-(4-AMINO-2-TERT-BUTYL-5-METHYL-PHENYLSULFANYL)-6-CYCLOPENTYL-4-HYDROXY-6-[2-(4-HYDROXY-PHENYL)-ETHYL]-5,6-DIHYDRO-PYRAN-2-ONE, Hepatitis C virus NS5B RNA polymerase | | Authors: | Love, R.A, Parge, H.E, Yu, X, Hickey, M.J, Diehl, W, Gao, J, Wriggers, H, Ekker, A, Wang, L, Thomson, J.A, Dragovich, P.S, Fuhrman, S.A. | | Deposit date: | 2003-03-18 | | Release date: | 2004-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic identification of a noncompetitive inhibitor binding site on the hepatitis C virus NS5B RNA polymerase enzyme.
J.Virol., 77, 2003
|
|
3Q22
 
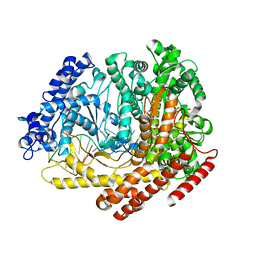 | |
8UO1
 
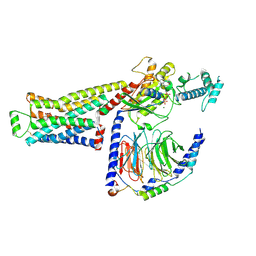 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class Q) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
6V9D
 
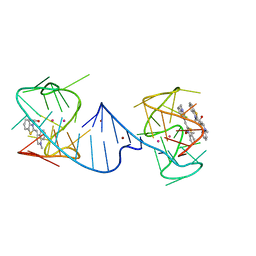 | |
6V9B
 
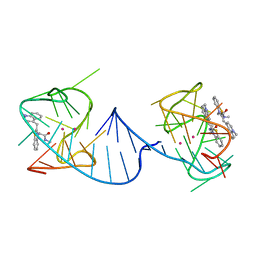 | |
3IVK
 
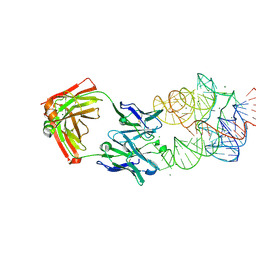 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
8GAP
 
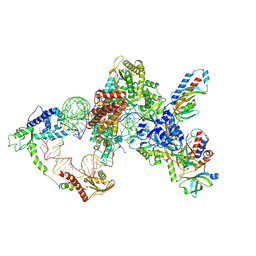 | | Structure of LARP7 protein p65-telomerase RNA complex in telomerase | | Descriptor: | Telomerase La-related protein p65, Telomerase RNA, Telomerase associated protein p50, ... | | Authors: | Wang, Y, He, Y, Wang, Y, Yang, Y, Singh, M, Eichhorn, C.D, Zhou, Z.H, Feigon, J. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of LARP7 Protein p65-telomerase RNA Complex in Telomerase Revealed by Cryo-EM and NMR.
J.Mol.Biol., 435, 2023
|
|
4QU6
 
 | |
7DMQ
 
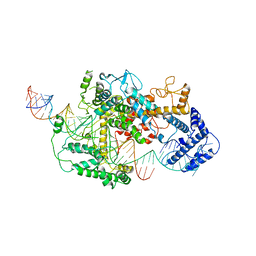 | | Cryo-EM structure of LshCas13a-crRNA-anti-tag RNA complex | | Descriptor: | Anti-tag target RNA, CRISPR RNA, CRISPR/Cas system Cas13a | | Authors: | Wang, B, Zhang, T, Ding, J, Patel, D.J, Yang, H. | | Deposit date: | 2020-12-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Mol.Cell, 81, 2021
|
|
483D
 
 | |
2B9Z
 
 | | Solution structure of FHV B2, a viral suppressor of RNAi | | Descriptor: | B2 protein | | Authors: | Lingel, A, Simon, B, Izaurralde, E, Sattler, M. | | Deposit date: | 2005-10-13 | | Release date: | 2005-11-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the flock house virus B2 protein, a viral suppressor of RNA interference, shows a novel mode of double-stranded RNA recognition.
Embo Rep., 6, 2005
|
|
3N6M
 
 | | Crystal structure of EV71 RdRp in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, NICKEL (II) ION, RNA-dependent RNA polymerase | | Authors: | Wu, Y, Lou, Z.Y, Miao, Y, Yu, Y, Rao, Z.H. | | Deposit date: | 2010-05-26 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of EV71 RNA-dependent RNA polymerase in complex with substrate and analogue provide a drug target against the hand-foot-and-mouth disease pandemic in China.
Protein Cell, 1, 2010
|
|
480D
 
 | |
3Q23
 
 | |
