7P0F
 
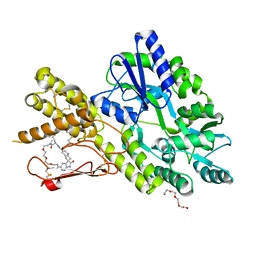 | | Crystal structure of a CGRP receptor ectodomain heterodimer bound to macrocyclic inhibitor HTL0028125 | | Descriptor: | (1S,10R,23E)-12-methyl-10-[(7-methyl-1H-indazol-5-yl)methyl]-15,18,21-trioxa-5,9,12,27,29-pentazapentacyclo[23.5.2.11,4.13,7.028,31]tetratriaconta-3(33),4,6,23,25(32),26,28(31)-heptaene-8,11,30-trione, Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M, Watson, S.P. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Novel Macrocyclic Antagonists of the Calcitonin Gene-Related Peptide Receptor: Design, Realization, and Structural Characterization of Protein-Ligand Complexes.
Acs Chem Neurosci, 13, 2022
|
|
6HRF
 
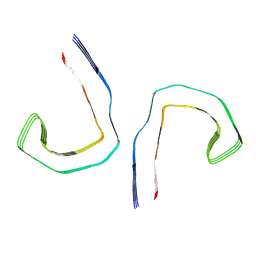 | | Straight filament from sporadic Alzheimer's disease brain | | Descriptor: | Microtubule-associated protein tau | | Authors: | Falcon, B, Zhang, W, Schweighauser, M, Murzin, A.G, Vidal, R, Garringer, H.J, Ghetti, B, Scheres, S.H.W, Goedert, M. | | Deposit date: | 2018-09-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Tau filaments from multiple cases of sporadic and inherited Alzheimer's disease adopt a common fold.
Acta Neuropathol., 136, 2018
|
|
6XRZ
 
 | | The 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Descriptor: | Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome | | Authors: | Zhang, K, Zheludev, I, Hagey, R, Wu, M, Haslecker, R, Hou, Y, Kretsch, R, Pintilie, G, Rangan, R, Kladwang, W, Li, S, Pham, E, Souibgui, C, Baric, R, Sheahan, T, Souza, V, Glenn, J, Chiu, W, Das, R. | | Deposit date: | 2020-07-14 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Biorxiv, 2020
|
|
6WJC
 
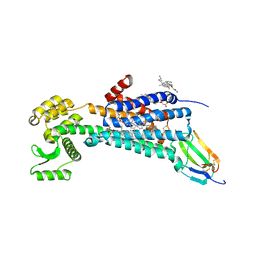 | | Muscarinic acetylcholine receptor 1 - muscarinic toxin 7 complex | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, ACETAMIDE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Kobilka, B.K. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and selectivity engineering of the M1muscarinic receptor toxin complex.
Science, 369, 2020
|
|
7PD9
 
 | |
7PCP
 
 | |
7PBY
 
 | |
6HAL
 
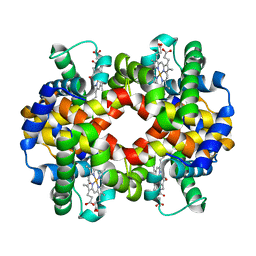 | | Human carbonmonoxy hemoglobin SFX dataset | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Doak, B, Gorel, A, Foucar, L, Barends, T.R.M, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C.M, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrell, D.A, Owen, R.L, Schlichting, I. | | Deposit date: | 2018-08-07 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7Q3V
 
 | |
3S8L
 
 | |
3S8O
 
 | |
4RGW
 
 | | Crystal Structure of a TAF1-TAF7 Complex in Human Transcription Factor IID | | Descriptor: | GLYCEROL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7 | | Authors: | Wang, H, Curran, E.C, Hinds, T.R, Wang, E.H, Zheng, N. | | Deposit date: | 2014-09-30 | | Release date: | 2014-12-03 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal structure of a TAF1-TAF7 complex in human transcription factor IID reveals a promoter binding module.
Cell Res., 24, 2014
|
|
3NB6
 
 | |
7ALC
 
 | | human GCH-GFRP stimulatory complex | | Descriptor: | GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, PHENYLALANINE, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.726 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7ALB
 
 | | human GCH-GFRP stimulatory complex 7-deaza-GTP bound | | Descriptor: | 7-deaza-GTP, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7PPN
 
 | | SHP2 catalytic domain in complex with CD28 (183-198) phosphopeptide (pTyr-191, p-Thr-195) | | Descriptor: | GLYCEROL, T-cell-specific surface glycoprotein CD28, Tyrosine-protein phosphatase non-receptor type 11,Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Sok, P, Zeke, A, Remenyi, A. | | Deposit date: | 2021-09-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the pSer/pThr dependent regulation of the SHP2 tyrosine phosphatase in insulin and CD28 signaling.
Nat Commun, 13, 2022
|
|
3S8R
 
 | | Crystal Structures of Glutaryl 7-Aminocephalosporanic Acid Acylase: Insight into Autoproteolytic Activation | | Descriptor: | GLYCEROL, Glutaryl-7-aminocephalosporanic-acid acylase | | Authors: | Kim, J.K, Yang, I.S, Park, S.S, Kim, K.H. | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of glutaryl 7-aminocephalosporanic acid acylase: insight into autoproteolytic activation.
Biochemistry, 42, 2003
|
|
6I2I
 
 | | Refined 13pf Hela Cell Tubulin microtubule (EML4-NTD decorated) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Atherton, J.M, Moores, C.A. | | Deposit date: | 2018-11-01 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression.
Sci.Signal., 12, 2019
|
|
4RZD
 
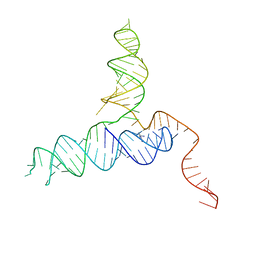 | | Crystal Structure of a PreQ1 Riboswitch | | Descriptor: | 7-DEAZA-7-AMINOMETHYL-GUANINE, PreQ1-III Riboswitch (Class 3) | | Authors: | Wedekind, J.E, Liberman, J.A, Salim, M. | | Deposit date: | 2014-12-20 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3U1H
 
 | |
6I41
 
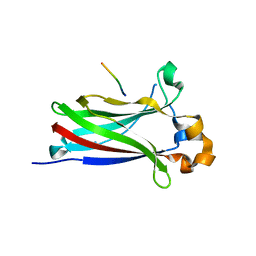 | |
8PBL
 
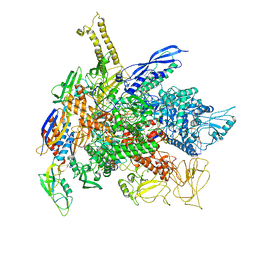 | |
3U0J
 
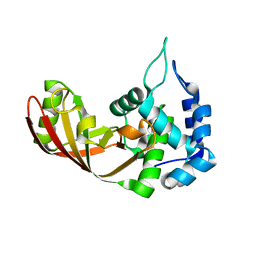 | |
4MGR
 
 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3UBP
 
 | | DIAMIDOPHOSPHATE INHIBITED BACILLUS PASTEURII UREASE | | Descriptor: | DIAMIDOPHOSPHATE, NICKEL (II) ION, PROTEIN (UREASE ALPHA SUBUNIT), ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Miletti, S, Mangani, S, Ciurli, S. | | Deposit date: | 1998-12-16 | | Release date: | 1999-12-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new proposal for urease mechanism based on the crystal structures of the native and inhibited enzyme from Bacillus pasteurii: why urea hydrolysis costs two nickels.
Structure Fold.Des., 7, 1999
|
|
