7OFU
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 3, 4-Dihydroxybenzoic acid, methyl ester | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Ewert, W, Werner, N, Falke, S, Guenther, S, Reinke, P, Sprenger, J, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Wolf, M, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFS
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with 4-(2-hydroxyethyl)phenol | | Descriptor: | 4-(2-hydroxyethyl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Ewert, W, Sprenger, J, Koua, F, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Hakanpaeae, J, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Wolf, M, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7OFT
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro in complex with p-hydroxybenzaldehyde | | Descriptor: | CHLORIDE ION, P-HYDROXYBENZALDEHYDE, POTASSIUM ION, ... | | Authors: | Srinivasan, V, Werner, N, Falke, S, Guenther, S, Reinke, P, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Ewert, W, Sprenger, J, Lieske, J, Koua, F, Ginn, H, Lane, T.J, Wolf, M, Yefanov, O, Gelisio, L, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Wahab, A, Choudary, I, Turk, D, Hinrichs, W, Chapman, H.N, Meents, A, Betzel, C. | | Deposit date: | 2021-05-05 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7M3I
 
 | |
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHK
 
 | | Crystal Structure of Apo/Unliganded SARS-CoV-2 Main Protease (Mpro) at 310 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9601 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJH
 
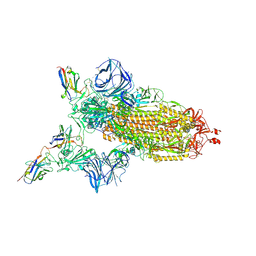 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJI
 
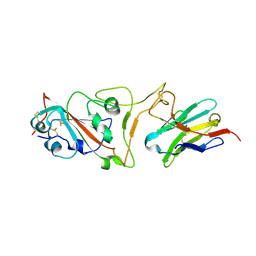 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to VH ab8 (focused refinement of RBD and VH ab8) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, VH ab8 | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MNG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor VBY-825 (Partial Occupancy) | | Descriptor: | (2R,3S)-N-cyclopropyl-3-{[(2R)-3-(cyclopropylmethanesulfonyl)-2-{[(1S)-2,2,2-trifluoro-1-(4-fluorophenyl)ethyl]amino}propanoyl]amino}-2-hydroxypentanamide (non-preferred name), 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Andi, B, Kumaran, D, Soares, A.S, Kreitler, D.F, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | Authors: | Yuan, M, Zhu, X, Wilson, I.A. | | Deposit date: | 2021-04-08 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.092 Å) | | Cite: | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
7MHL
 
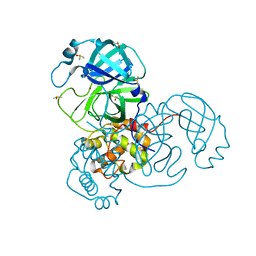 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJN
 
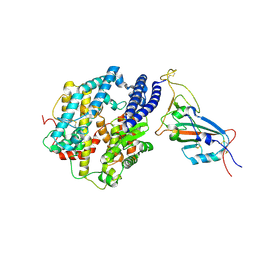 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MMO
 
 | | LY-CoV1404 neutralizing antibody against SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LY-CoV1404 Fab heavy chain, LY-CoV1404 Fab light chain, ... | | Authors: | Hendle, J, Pustilnik, A, Sauder, J.M, Coleman, K.A, Boyles, J.S, Dickinson, C.D. | | Deposit date: | 2021-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.427 Å) | | Cite: | LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants.
Biorxiv, 2022
|
|
7MHJ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K and High Humidity | | Descriptor: | 3C-like proteinase, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0005 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJJ
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MHH
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 277 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1908 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJL
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (focused refinement of RBD and Fab ab1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, Fab ab1 Light Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJK
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to Fab ab1 (class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab ab1 Heavy Chain, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MHI
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 298 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MJG
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7MJM
 
 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
7B27
 
 | |
7LCQ
 
 | | N-terminal finger stabilizes feline drug GC376 in coronavirus 3CL protease | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Khan, M.B, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-Terminal Finger Stabilizes the S1 Pocket for the Reversible Feline Drug GC376 in the SARS-CoV-2 M pro Dimer.
J.Mol.Biol., 433, 2021
|
|
