9ICD
 
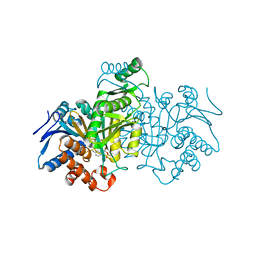 | | CATALYTIC MECHANISM OF NADP+-DEPENDENT ISOCITRATE DEHYDROGENASE: IMPLICATIONS FROM THE STRUCTURES OF MAGNESIUM-ISOCITRATE AND NADP+ COMPLEXES | | Descriptor: | ISOCITRATE DEHYDROGENASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hurley, J.H, Dean, A.M, Koshland Jr, D.E, Stroud, R.M. | | Deposit date: | 1991-07-29 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic mechanism of NADP(+)-dependent isocitrate dehydrogenase: implications from the structures of magnesium-isocitrate and NADP+ complexes.
Biochemistry, 30, 1991
|
|
9ICC
 
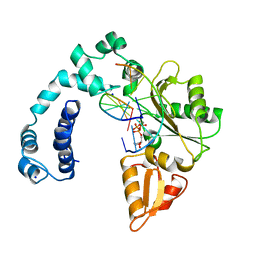 | |
9ICB
 
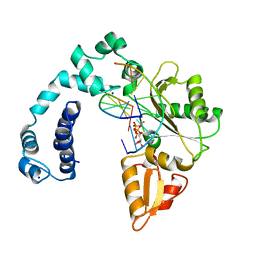 | |
9ICA
 
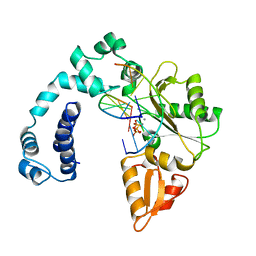 | |
9HVP
 
 | | Design, activity and 2.8 Angstroms crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease | | Descriptor: | HIV-1 Protease, benzyl [(1R,4S,6S,9R)-4,6-dibenzyl-5-hydroxy-1,9-bis(1-methylethyl)-2,8,11-trioxo-13-phenyl-12-oxa-3,7,10-triazatridec-1-yl]carbamate | | Authors: | Neidhart, D.J, Erickson, J. | | Deposit date: | 1990-11-06 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, activity, and 2.8 A crystal structure of a C2 symmetric inhibitor complexed to HIV-1 protease.
Science, 249, 1990
|
|
9GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH S-HEXYL GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE S-TRANSFERASE P1-1, S-HEXYLGLUTATHIONE, ... | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
9GPB
 
 | |
9GAF
 
 | |
9GAC
 
 | |
9GAA
 
 | |
9G2A
 
 | | Staphylococcus aureus MazF in complex with nanobody 4 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Endoribonuclease MazF, ... | | Authors: | Prolic-Kalinsek, M, Zorzini, V, Haesaerts, S, Loris, R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.046594 Å) | | Cite: | Staphylococcus aureus MazF in complex with nanobody 4
To Be Published
|
|
9G0K
 
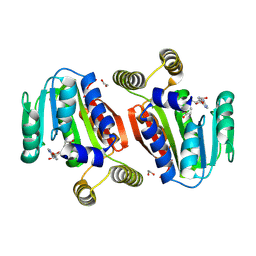 | | CysG(N-16) in complex with SAH (metal free, cocrystallization in presence of EDTA) from Kitasatospora cystarginea | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Kuttenlochner, W, Beller, P, Kaysser, L, Groll, M. | | Deposit date: | 2024-07-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Deciphering the SAM- and Metal-Dependent Mechanism of O-Methyltransferases in Cystargolide and Belactosin Biosynthesis: A Structure-Activity Relationship Study
to be published
|
|
9FZK
 
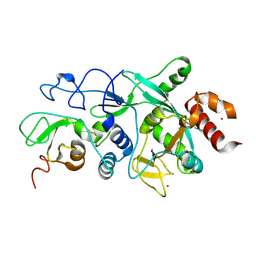 | |
9FZD
 
 | |
9FZC
 
 | |
9FZ4
 
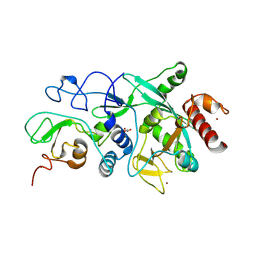 | |
9FYP
 
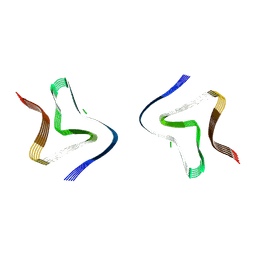 | | Cryo EM structure of the type 3B polymorph of alpha-synuclein at low pH. | | Descriptor: | Alpha-synuclein, CHLORIDE ION | | Authors: | Frey, L, Qureshi, B.M, Kwiatkowski, W, Rhyner, D, Greenwald, J, Riek, R. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | On the pH-dependence of alpha-synuclein amyloid polymorphism and the role of secondary nucleation in seed-based amyloid propagation
Elife, 2024
|
|
9FYB
 
 | | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Pandian, R, Jeje, O.A, Sayed, Y, Achilonu, I.A. | | Deposit date: | 2024-07-03 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural Insights into the NMN Complex of Nicotinate Nucleotide Adenylyltransferase from Enterococcus faecium via Co-Crystallization Studies
To be published
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
9FWC
 
 | | Coxsackievirus B3 3C protease in C121 spacegroup | | Descriptor: | Genome polyprotein | | Authors: | Fairhead, M, Lithgo, R.M, MacLean, E.M, Bowesman-Jones, H, Aschenbrenner, J.C, Balcomb, B.H, Capkin, E, Chandran, A.V, Godoy, A.S, Marples, P.G, Fearon, D, von Delft, F, Koekemoer, L. | | Deposit date: | 2024-06-28 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Coxsackievirus B3 3C protease in C121 spacegroup
To Be Published
|
|
9FW2
 
 | |
9FVT
 
 | |
9FVO
 
 | | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Endothiapepsin, ... | | Authors: | Falke, S, Senst, J.M, Guenther, S, Meents, A. | | Deposit date: | 2024-06-27 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The HIV protease inhibitor amprenavir binding to the active site of Cryphonectria parasitica endothiapepsin
To Be Published
|
|
9FT1
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 13 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
