3CHR
 
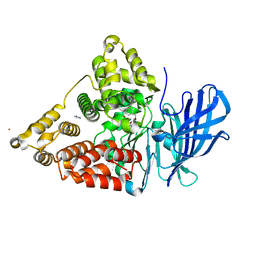 | | Crystal structure of leukotriene A4 hydrolase in complex with 4-amino-N-[4-(phenylmethoxy)phenyl]-butanamide | | Descriptor: | 4-amino-N-[4-(benzyloxy)phenyl]butanamide, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
1Z5M
 
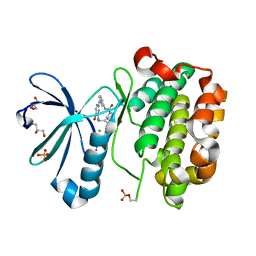 | |
2JG8
 
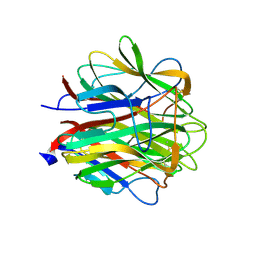 | | Crystallographic structure of human C1q globular heads complexed to phosphatidyl-serine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1q subcomponent subunit A, ... | | Authors: | Paidassi, H, Tacnet-Delorme, P, Garlatti, V, Darnault, C, Ghebrehiwet, B, Gaboriaud, C, Arlaud, G.J, Frachet, P. | | Deposit date: | 2007-02-09 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | C1Q Binds Phosphatidylserine and Likely Acts as a Multiligand-Bridging Molecule in Apoptotic Cell Recognition.
J.Immunol., 180, 2008
|
|
3CZD
 
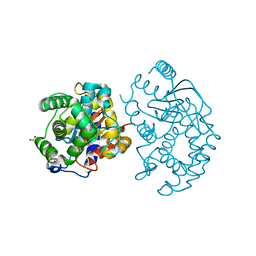 | | Crystal structure of human glutaminase in complex with L-glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutaminase kidney isoform, ... | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2JSI
 
 | | 11-23 obestatin fragment in DPC/SDS micellar solution | | Descriptor: | Appetite-regulating hormone, Obestatin | | Authors: | D'Ursi, A.M, Scrima, M, Esposito, C, Campiglia, P. | | Deposit date: | 2007-07-05 | | Release date: | 2008-10-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Obestatin conformational features: a strategy to unveil obestatin's biological role?
Biochem.Biophys.Res.Commun., 363, 2007
|
|
1ZF1
 
 | | CCC A-DNA | | Descriptor: | 5'-D(*CP*CP*GP*GP*GP*CP*CP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFA
 
 | | GGA Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*TP*CP*CP*GP*GP*AP*GP*G)-3', CALCIUM ION, SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3L6B
 
 | | X-ray crystal structure of human serine racemase in complex with malonate a potent inhibitor | | Descriptor: | MALONATE ION, MANGANESE (II) ION, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Smith, M.A, Barker, J, Mack, V, Ebneth, A, Moraes, I, Felicetti, B, Cesura, A. | | Deposit date: | 2009-12-23 | | Release date: | 2010-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of mammalian serine racemase: evidence for conformational changes upon inhibitor binding.
J.Biol.Chem., 285, 2010
|
|
1ZF8
 
 | | GGT Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*GP*GP*TP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3CEQ
 
 | | The TPR domain of Human Kinesin Light Chain 2 (hKLC2) | | Descriptor: | Kinesin light chain 2 | | Authors: | Zhu, H, Shen, Y, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-08-12 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The TPR domain of Human Kinesin Light Chain 2 (hKLC2)
To be Published
|
|
1ZHB
 
 | | Crystal Structure Of The Murine Class I Major Histocompatibility Complex Of H-2Db, B2-Microglobulin, and a 9-Residue Peptide Derived from rat dopamine beta-monooxigenase | | Descriptor: | 9-mer peptide from Dopamine beta-monooxygenase, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | Authors: | Sandalova, T, Michaelsson, J, Harris, R.A, Odeberg, J, Schneider, G, Karre, K, Achour, A. | | Deposit date: | 2005-04-25 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
J.Biol.Chem., 280, 2005
|
|
3QG7
 
 | | Structural Basis for Ligand Recognition and Discrimination of a Quorum Quenching Antibody | | Descriptor: | AP4-24H11 Antibody Heavy Chain, AP4-24H11 Antibody Light Chain, HEXAETHYLENE GLYCOL, ... | | Authors: | Kirchdoerfer, R.K, Kaufmann, G.F, Janda, J.D, Wilson, I.A. | | Deposit date: | 2011-01-24 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Basis for Ligand Recognition and Discrimination of a Quorum-quenching Antibody.
J.Biol.Chem., 286, 2011
|
|
3CHS
 
 | | Crystal structure of leukotriene A4 hydrolase in complex with (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid | | Descriptor: | (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
2LK1
 
 | |
3AWN
 
 | | Crystal structure of D-serine dehydratase from chicken kidney (EDTA treated) | | Descriptor: | D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AWO
 
 | | Crystal structure of D-serine dehydratase in complex with D-serine from chicken kidney (EDTA-treated) | | Descriptor: | D-SERINE, D-serine dehydratase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Tanaka, H, Senda, M, Venugopalan, N, Yamamoto, A, Senda, T, Ishida, T, Horiike, K. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of a zinc-dependent D-serine dehydratase from chicken kidney.
J.Biol.Chem., 286, 2011
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
1Z1L
 
 | | The Crystal Structure of the Phosphodiesterase 2A Catalytic Domain | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, ZINC ION, ... | | Authors: | Ding, Y.H, Kohls, D, Low, C. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Determinants for Inhibitor Specificity and Selectivity in PDE2A Using the Wheat Germ in Vitro Translation System.
Biochemistry, 44, 2005
|
|
1ZF9
 
 | | GGG Duplex A-DNA | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1ZFM
 
 | | AGC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*TP*AP*GP*CP*GP*G)-3' | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A9Y
 
 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine | | Descriptor: | 6N-DIMETHYLADENOSINE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | Deposit date: | 2005-07-12 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2GTK
 
 | | Structure-based Design of Indole Propionic Acids as Novel PPARag CO-Agonists | | Descriptor: | (2S)-3-(1-{[2-(2-CHLOROPHENYL)-5-METHYL-1,3-OXAZOL-4-YL]METHYL}-1H-INDOL-5-YL)-2-ETHOXYPROPANOIC ACID, Decamer from Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Kuhn, B, Hilpert, H, Benz, J, Binggeli, A, Grether, U, Humm, R, Maerki, H.-P, Meyer, M, Mohr, P. | | Deposit date: | 2006-04-28 | | Release date: | 2006-09-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of indole propionic acids as novel PPARalpha/gamma co-agonists
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3D0E
 
 | | Crystal structure of human Akt2 in complex with GSK690693 | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3-yn-2-ol, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Smallwood, A. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-21 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase.
J.Med.Chem., 51, 2008
|
|
3CST
 
 | |
2GYV
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
